Heat shock proteins (HSPs) are a specific group of proteins that can be induced by heat shock (or stress) in a wide range of organisms including bacteria, plants, insects, mammals, and humans (Lemaux et al., 1978; Yamamori et al., 1978; Kelley and Schlesinger, 1978 ; Miller et al., 1979 ; McAlister and Finkelstein, 1980 ; Johnston et al., 1980 ; Manjunatha et al., 2010 ; Routray et al., 2013). As the name suggests, HSPs are induced in cells, tissues, or individuals exposed to sublethal heat shock. HSPs were first discovered in 1962 by Ritossa . It was reported that when Drosophila salivary gland cells were exposed to 37℃ for 30 min then returned to their normal temperature of 25℃ for recovery, “puffing” of the corresponding genes was induced in the chromosomes of the recovering cells (Ritossa, 1962). In the 1970s, researchers found that this phenomenon was accompanied by an increase in the expression of proteins with molecular masses of 26 and 70 kilodalton (kDa) (Tissieres et al., 1974). These proteins were named “heat shock proteins”. Since these discoveries were made, a large number of proteins, referred to as HSPs, have been reported.
In addition to heat shock, the synthesis of HSPs can be induced by a variety of chemical and environmental stimuli. Exposure of cells to heavy metals (Levinson et al., 1980), amino acid analogs (Kelley and Schlesinger, 1978; Li and Laszlo, 1985), glucose analogs (Pouyssegur et al., 1977), Ca2+ increasing agents (Ding et al., 1996), ischemia, sodium arsenite (Johnson et al., 1980), microbial infections, nitric oxide, hormones, and antibiotics also induce the expression of HSPs. HSPs have been shown to perform multiple functions in cellular processes. Notably, HSPs function as molecular chaperones (Ellis, 1987) to facilitate proper synthesis and folding of proteins throughout the cell (Walter and Buchner, 2002; Martin and Ulrich, 1997). They also stabilize and assist in the trafficking of nascent peptides during normal growth (Hartl et al., 1992). Under stressful conditions such as heat shock or hypoxia (Hammerer, 2001), increased expression of HSPs protects the cell by stabilizing unfolded or misfolded peptides and by allowing the cell time to repair or re-synthesize damaged proteins (Saibil, 2000). HSPs have been shown to participate in protein assembly (Welch, 1991), secretion (Wild et al., 1992), and degradation as well as cellular homeostasis (Mallouk et al., 1999), and the regulation of transcription factors and protein kinases (Sonya et al., 2003). Additionally, HSPs have appeared to serve a protective role in cardiovascular health by aiding (Benjamin and McMillan, 1998) smooth muscle relaxation (McLemore et al., 2005), preventing platelet aggregation, improving cardiac myocyte function, and preventing apoptosis after ischemic injury (Fan, 2005). Extracellular and membrane bound HSPs are involved in binding antigens and presenting them to the immune system (Nishikawa et al., 2008); therefore, HSPs are useful as immunologic adjuvants in boosting the response to a vaccine (Bendz et al., 2007). It has been speculated that HSPs may be useful for increasing the effectiveness of cancer vaccines (Binder, 2008).
Recent studies on HSPs have implicated their role in physiological, biochemical, molecular, biological, ecological, evolutionary, and clinical research. These studies have led to cloning and characterization of genes encoding HSPs in many organisms including the entomopathogenic fungus in our present study. Until now, a great number of variable species of HSPs including small HSPs with molecular weights ranging from 15-40 kDa and many types of HSPs with molecular weights over 40 kDa have been studied; however, HSP88 is rarely investigated (Schlesinger, 1990; Sorensen et al., 2003; Davis, 2004; Burnie et al., 2006; Bendz et al., 2007; Tsan and Gao, 2009; Manjunatha et al., 2010; Shim et al., 2012; Routray et al., 2013). Although the amino acid sequence of the HSP88 has been reported from a fungal species (Plesofsky-Vig and Brambl, 1998), there are few reports regarding the full-length cDNA sequence. This is the first report, to our knowledge, about the HSP88 from Paecilomyces tenuipes Jocheon-1 (P. tenuipes Jocheon-1), an entomopathogenic fungus also known as the vegetable wasp, and plant worm. P. tenuipes Jocheon-1 is an ecotype of the corresponding species. Dongchoonghacho, a kind of silkworm mushroom produced from the entomopathogenic fungus of P. tenuipes, has been known as a mysterious ancient medicine in Asian countries, particularly in China, Korea, and Japan. This paper describes the molecular cloning of HSP88 cDNA from P. tenuipes Jocheon-1, the expression of cDNA, the deduced amino acid sequence of the protein, and the influence of the heat shock treatment on the corresponding mRNA expression levels.
The entomopathogenic fungal strain, P. tenuipes Jocheon-1, was used in this study. This strain was collected from the mountainous region of Miryang, South Korea and was artificially cultured in egg yolk medium as described previously (Lee et al., 2006).
Total RNA was isolated from the fruiting body of P. tenuipes Jocheon-1 using the SV Total RNA Isolation System (Promega ). mRNA was isolated using the oligotex mRNA midi kit (Qiagen, Korea Ltd). cDNA was synthesized using the Uni-ZAP cDNA synthesis kit (Stratagene) according to the manufacturer’s instruction. cDNA was ligated into the EcoRI and XhoI sites of the Uni-ZAP XR vector and packaged in the high efficiency system GigapackIII Gold packaging extract (Stratagene).
The Uni-ZAP XR lambda cDNA library clones were converted to the pBluescript SK(-) phagemid by in vivo excision as described by the manufacturer. Escherichia coli (E. coli) SOLR cells were infected with the phagemid, plated onto LB ampicillin agar plates, and incubated at 37℃ overnight. Colonies appearing on the plate with the cDNA insert were selected for plasmid isolation. Plasmid DNA isolation was extracted using Accuprep plasmid mini extraction kit (Bioneer). Plasmids were then subjected to sequence analysis. P. tenuipes Jocheon-1 ESTs were compared with the BLASTX of the NCBI database to identify the homologous genes.
According to the P. tenuipes Jocheon-1 UniZAP cDNA library sequencing results, one clone with the insert of 1,444 basepairs (bp) showed higher similarity to the previously reported fungal HSP88. This clone contained the complete 3′-end with a poly(A) tail, but it was incomplete at the 5′-end. In order to obtain the full-length sequence of this predicted HSP88 cDNA, 5′ RACE PCR was performed using the Marathon cDNA amplification kit (Clontech) according to the manufacturer’s protocol. In order to guarantee the specificity of the PCR product, Nested PCR was performed using the following primers: Gene specific primer (GSP), nested gene specific primer (NGSP), adaptor primer 1 (AP1), and adaptor primer 2 (AP2) were supplied with the marathon cDNA amplification kit (Clontech). Amplifications were performed with a MyCycler Thermal Cycler (BioRad). The two specific primers were designed based on this partial sequence information. In the 5′ RACE reaction, adaptor-ligated cDNA was generated from poly(A) RNA isolated from the fruiting body of P. tenuipes Jocheon-1. The first PCR amplification was carried out using lower GSP1 (5′-TGAATGTTAACACGGGTCTTAAGA-3′) and AP1 (5′-CCATCCTAATACGACTCAC-TATAGGGC-3′). PCR cycling conditions were as follows: 33 cycles at 94℃ for 1 min, 60℃ for 1 min, and 72℃ for 2.5 min followed by a terminal extension cycle at 72℃ for 7 min. The second PCR amplification was carried out using upper GSP2 (5′-TGAATGTTAACACGGGTCTTAAGA-3′) and AP2 (5′-ACTCACTATAGGGCTCGAGCGGC-3′). The PCR cycling conditions were the same as with the first PCR cycling conditions except that the annealing temperature was 63℃. The PCR product was analyzed with a 1% agarose gel electrophoresis, and the band of approximately 1.4 kilobasepairs (kbp ) was excised from the gel. The band was then purified with the AccuPrep Gel purification kit (Bioneer) and cloned to the pGEM-T Easy vector (Promega, USA) following the manufacturer’s instruction. Blue-white screening of HIT competent DH5a cells was used to identify potential transformants. Positive clones were selected and sequenced with an Applied Biosystem 3730XI DNA Analyzer (USA). The 5′ region sequencing result was combined with the poly(A) tail connected to the 3′ region sequence to make the predicted full-length cDNA sequence.
To isolate the full-length clone, the Marathon cDNA library of P. tenuipes Jocheon-1 was screened with the HSP forward primer (PtHSP-F 5′-AGCAACCATGAGTGTCGTCGGTG-3′) and the HSP reverse primer (PtHSP-R 5′-CGTGATATCTATTTGGAATCGGC-3′). These were designed based on the combined sequence information of the 5′-end sequence and the previously obtained partial sequence. Samples were run on a thermal cycle (BioRad) using PCR PreMix (Bioneer) under the following conditions: 35 cycles at 94℃C for 1 min, 53℃ for 1 min, and 72℃ for 2.5 min. The amplified fragment of approximately 2.1 kbp was purified and cloned to the pGEM-T Easy Vector that was then transformed to HIT competent DH5a cells (RH617, RBC). The plasmid was isolated using a plasmid mini extraction kit and sequenced with an Applied Biosystems 3730xl DNA Analyzer (USA).
The amino acid sequences of other reported HSP88s were retrieved from the GenBank database provided by the NCBI. The sequences were aligned in clustal W (Thompson et al., 1994). Molecular evolutionary genetics analysis (MEGA) software was used for statistical and phylogenetic analysis (Kumar et al., 1994). The phylogenetic tree was constructed with the neighbor joining method (Saitou and Nei, 1987).
The 2,139-bp open reading frame (ORF) of HSP88 cDNA from P. tenuipes Jocheon-1 was subcloned between the SbfI and BglII sites of pBacPAK9 (Clontech) to produce a baculovirus transfer vector pBacPAK9-PtHSP88. In the transfer vector, the P. tenuipes Jocheon-1 HSP88 cDNA was under the control of the polyhedrin promoter of Autographa californica nuclear polyhedrosis virus (AcNPV).
Spodoptera fruigiperda IPLB Sf21-AE (Vaughn et al., 1977) clone9 (Sf9) cells were maintained at 27℃ in TC100 medium (Sigma) supplemented with 10% fetal bovine serum (FBS, Sigma). Cells were cultured as described by standard methods (O'Reilly et al., 1992). Wild type AcNPV and recombinant AcNPV were propagated in Sf9 cells. The titer was expressed as plaque forming units (PFU) per mL (O'Reilly et al., 1992).
Thirty-five milliliter cell culture dishes were seeded with 1.0-1.5 × 106 cells and incubated at 27℃ for 1 h to allow cell attachment. One microgram of BacPAK6 viral DNA (Clontech), 5 μg of pBacPAK9-PtHSP88 in 20 mM HEPES buffer, and sterile water were mixed in a polystyrene tube for a total volume of 50 μL. Fifty μL of 100 μg/mL Lipofectin (Gibco Brl Life Technologies) were gently mixed with the DNA solution and the mixture was incubated at the room temperature for 30 min. The cells were washed twice with 2 mL serum free TC100 medium and fed with 1.5 mL serum free TC100 medium. The Lipofectin-DNA complexes were added in drops to the medium covering the cells while each dish was gently swirled. After incubation at 27℃ for 5 h, TC100 medium containing antibiotics and 10% FBS was added to each dish and returned to incubation at 27℃. After 3 d, the supernatant was harvested, clarified by centrifugation at 2,000 rpm for 5 min, and stored at 4℃ until use. Recombinant AcNPV was plaque purified as described by O'Reilly et al. (1992).
Sf9 insect cells were mock-infected, infected with wild type Autographa californica nuclear polyhedrosis virus (AcNPV), or recombinant AcNPVs at a multiplicity of infection of 106 in 35 mm dishes. After incubation at 27°C for 3 d, cells were harvested. For SDS-PAGE of cell lysates, uninfected cells and virus infected Sf9 cells were washed twice with phosphate buffered saline (PBS), mixed with protein sample buffer, and boiled . Total cellular lysates were then subjected to 10% SDS-PAGE.
The normal temperature for cultivation of P. tenuipes Jocheon-1 in the developmental stage of fruit body is approximately 20℃. In order to investigate the effects of heat shock treatments on the expression level of HSP88 mRNA in the early fruit body developmental stage, P. tenuipes Jocheon-1 were kept at 25℃ and 30℃ for different amounts of time, 8 h, 16 h, or 24 h.
qPCR was used to measure the relative expression levels of HSP88 during the developmental stages of mycelium, early fruit body, middle fruit body, and late fruit body, respectively. qPCR has also previously been performed to analyze the effect of heat treatment on the relative expression level of HSP88 when P. tenuipes Jocheon-1 was in early fruit body development. One μg of total RNA was used for cDNA synthesis. cDNA was synthesized using the iScript cDNA synthesis kit (Biorad). qPCR was performed using an iQ5™ multicolor real time PCR detection system (BioRad) and sensimix SYBR kit (Bioline, according to the manufacturer’s instructions.
Two HSP88 specific primers (RT-HSP-F ; 5′-GCCAGTGAAGAGGAGAAG-3′ and RT-HSP-R; 5′-GCGATGTAGACACCCTTT-3′) were used to amplify a PCR product of 101 bp. Two GAPDH primers (GAPDH-F; 5′-CACTCCTACACCGCTACC-3′, GAPDG-R; 5′-GAATGACCTTGCCGACAG-3′) were used to amplify a GAPDH gene fragment of 130 bp. GAPDH primers were based on sequences from GenBank Accession number GU997101. GAPDH was used as an internal control for qPCR. The qPCR amplifications were carried out in total volume of 20 μL containing 10 μL of SYBR, 1 μL of cDNA, 0.5 μL of each forward and reverse primer (5 pmol final concentration), and 8 μL of DEPC water.
Reactions were initiated with a denaturation step at 95℃ for 10 min, followed by 40 cycles at 95℃ for 15 s, 55℃ for 30 s, 72℃ for 20 s, and a dissociation curve at 95℃ for 60 s, 55℃ for 60 s, and 60℃ for 10 s. Two independent biological replicates were carried out . Melting curve analysis of amplification products was performed at the end of each PCR to confirm that only one product was amplified and detected. The comparative cycle threshold (Ct) method was used to analyze the expression level of HSP88 .
A cDNA library was constructed using fruiting bodies of P. tenuipes Jocheon-1. One thousand clones were randomly selected. Clones that were of low quality, too short (<700 bp), or had vector contamination were not included. Seven hundred and forty one ESTs were generated by a partial sequencing from the cDNA library. Of the 741 ESTs, 305 showed homology to genes identified and deposited in the NCBI database, 261 matched to unknown or unclassified functional genes, and 175 displayed similarity with an E-value that was lower than the adopted standard value of 10-5 in the process of functional annotation. According to the EST results of the cDNA library, one clone with an insert of 1,444 bp showed high homology to the previously reported HSP88 gene. The sequence contained the complete 3′-end with a poly(A) tail, but it was not complete at the 5′-end. Accordingly, the marathon 5′ RACE PCR amplification method was used to determine the unknown 5′-end sequence as described previously. The PCR product was excised from the gel, purified with a gel extraction kit (Promega), and subcloned into the pGEM-T Easy vector for sequence analysis.
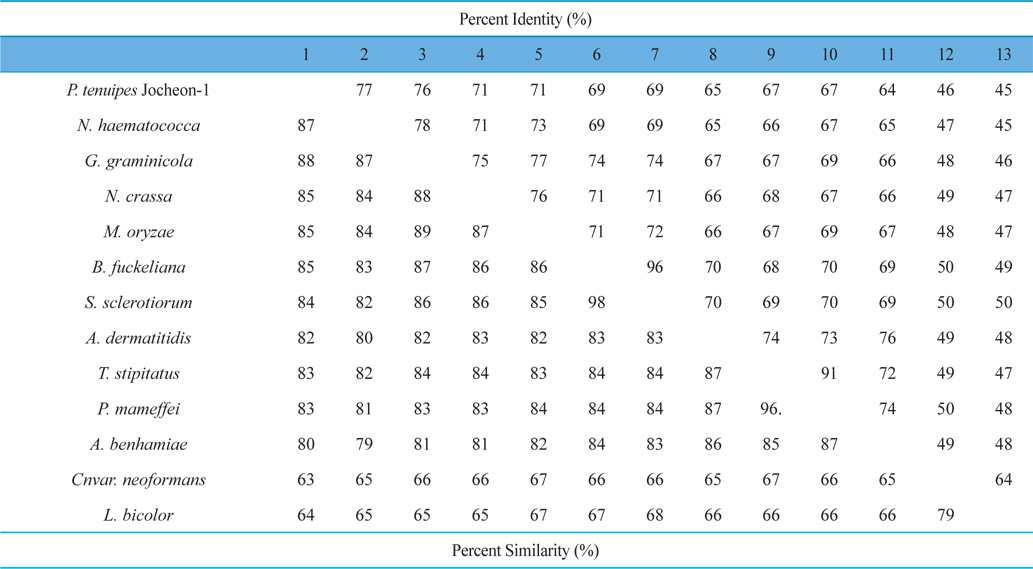
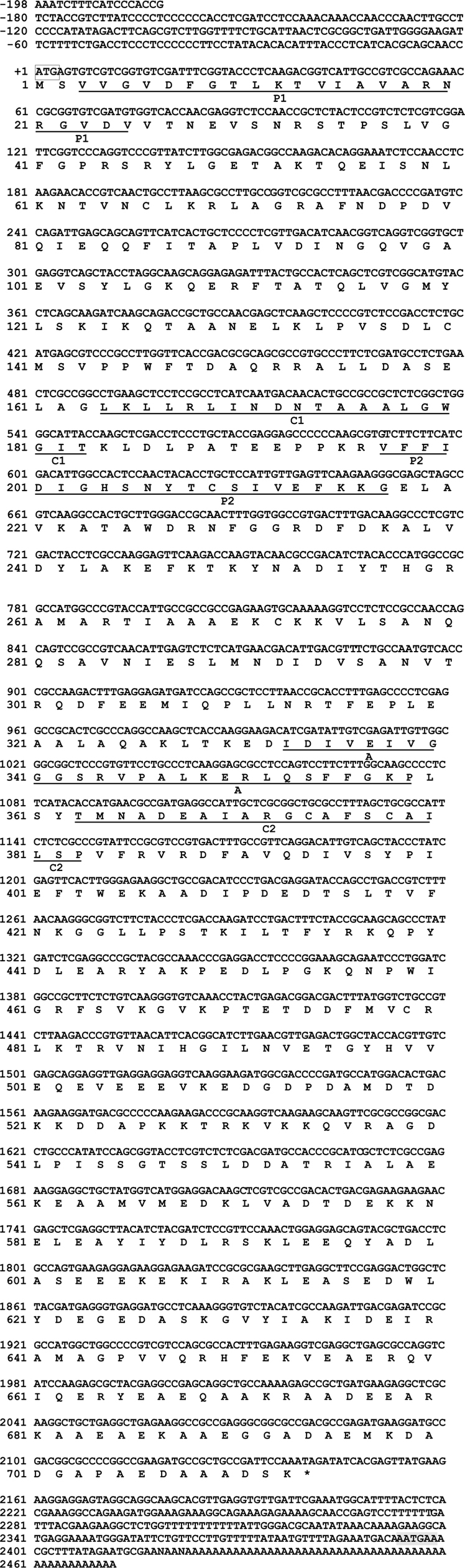
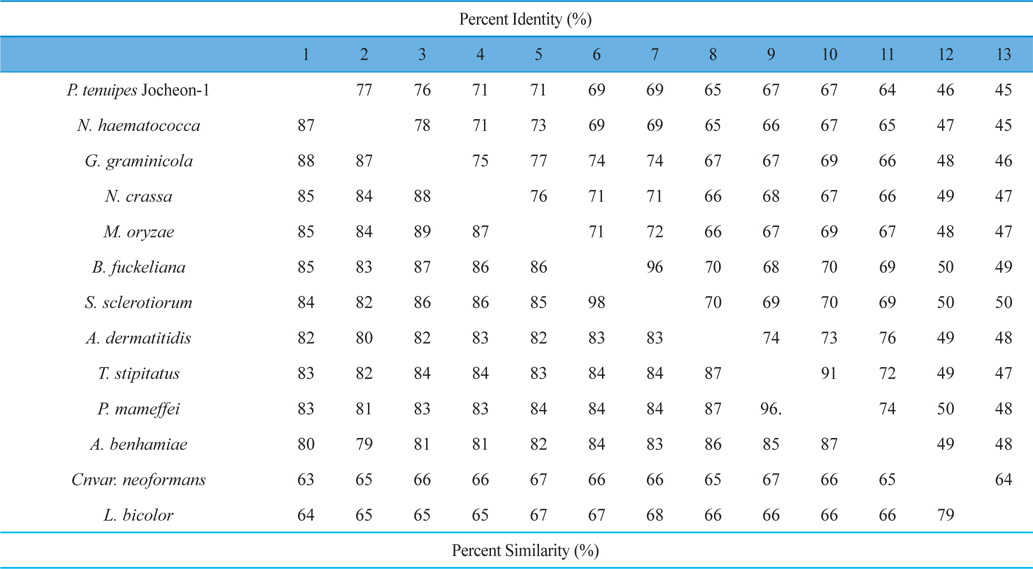
The DNA fragment of approximately 2.1 kbp that amplified using the primer pairs PtHSP-F and PtHSP-R was excised and subcloned into the pGEM-T Easy vector for sequencing. The nucleotide and deduced amino acid sequences of HSP88 cDNA are represented in Fig. 1. The cDNA contained an ORF of 2,139-bp encoding 713 amino acid residues. From the nucleotide and deduced amino acid sequence of the cDNA, five sequence motifs named phosphate1, connect1, phosphate2, adenosine, and connect2, which involved in ATP binding and interdomain coupling (Buchberger et al., 1994), have been identified (Fig. 1). Alignment of deduced amino acid sequences of P. tenuipes Jocheon-1 HSP88 cDNA with other fungal HSP88s is represented in Fig. 2. This data demonstrates that there are variable identities in the amino acid sequences of the homologous genes ranging from 45-77% (Fig. 2 and Table 1). Further comparison of the P. tenuipes Jocheon-1 HSP88 s equence with the ATPase domain of HSP70 , the ATP-binding cell division protein FtsA of E. coli (Bork et al., 1992), and the fungal HSP88 of Neurospora crassa (N. crassa) (Plesofsky-Vig and Brambl, 1998) revealed that P. tenuipes Jocheon-1 HSP88 also contains the conserved ATPase domain at the N-terminal region (Fig. 1).
[Fig. 1.] The nucleotide sequence, deduced amino acid sequence, and ATPase domain of the P. tenuipes Jocheon-1 HSP88 cDNA. The start codon, ATG, is boxed, and the termination codon is denoted with the asterisk. The poly(A) signal sequence is shown with the shadow mark. P1, phosphate1; P2, phosphate2; C1, connect1 (interdomain hinge); C2, connect2 (interdomain hinge); A, adenosine. This cDNA sequence has been provided with the accession number GU983958 from GenBank.
[Fig. 2.] Alignment of the amino acid sequences of P. tenuipes Jocheon-1 HSP88 with other known fungal HSP88s. GenBank accession numbers are: P. tenuipes Jocheon-1 (GU983958), G. graminicola (EFQ27583), N. haematococca (EEU34379), M. oryzae (EDJ96308), N. crassa (EDN96960), S. Sclerotio (EEP78775), B. fuckeliana (EDN30753), T. stipitatu (EED21621), P. marneffei (EEA25736), A. dermatitid (EEQ88845), A. benhamiae (EDN02919), Cn var. neoformans (AAW46676), and L. bicolor (EDR11689).
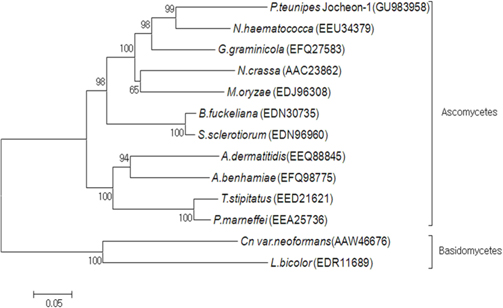
A phylogenetic analysis using the deduced amino acid sequences of known fungal HSP88 genes revealed that it is divided into two separate clades, ascomycetes and basidiomycetes (Fig. 3). Within the ascomycetes fungal clade, P. tenuipes Jocheon-1 and Nectria haematococca (N. haematococca) formed a subgroup, P. tenuipes Jocheon-1 HSP88 showed the highest protein sequence identity (77%) to N. haematococca. The identity of P. tenuipes Jochoen-1 to other known HSP88s was 45-76% (Table1).
[Fig. 3.] Phylogenetic tree of aligned amino acid sequences of the P. tenuipes Jocheon-1 HSP88 and the other known fungal HSP88 sequences. The accession numbers of the sequences in the GenBank are as follows: P. tenuipes Jocheon-1 (GU983958), N. hamematococca (EEU34379), G. graminicola (EFQ27583), N. crassa (AAC23862), M. oryzae (EDJ96308), B. fuckeliana (EDN30735), S. sclerotiorum (EDN96960), A. dermatitidis (EEQ88845), A. benhamiae (EFQ98775), T. stipitatus (EED21621), P. marneffei (EEA25763), Cn var. neoformans (AAW46676), and L. bicolor (EDR11689).
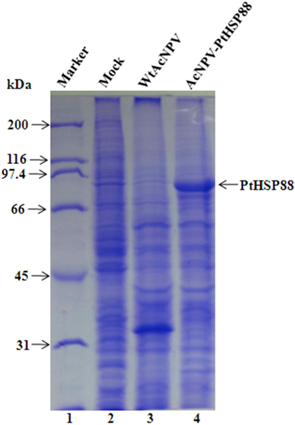
To assess the P. tenuipes Jocheon-1 HSP88 gene, the ORF of HSP88 cDNA, which was comprised of 2,139 nucleotides, was inserted into a baculovirus transfer vector. The transfer vector pBacPAK9-PtHSP88 was constructed by inserting P. tenuipes Jocheon-1 HSP88 cDNA under the control of the AcNPV polyhedrin promoter of pBacPAK9. In order to confirm the correct insertion of the HSP88 ORF fragment, the transfer vector pBacPAK9-PtHSP88 was double digested with restriction enzymes SbfI and BglII. The baculovirus transfer vector was used to generate a recombinant virus expressing P. tenuipes Jocheon-1 HSP88. The recombinant AcNPV (AcNPV-PtHSP88) was produced in insect Sf9 cells by co-transfection with wild type AcNPV DNA and the transfer vector. To examine expression, the protein synthesized in Sf9 cells infected with the recombinant virus was analyzed by SDS-PAGE. The recombinant P. tenuipes Jocheon-1 HSP88 expressed by the P. tenuipes Jocheon-1 HSP88 cDNA was approximately 88 kDa as determined by the gel from the cells infected with the recombinant virus but was not detected from the cells infected with the wild type AcNPV or mock-infected cells (Fig. 4).
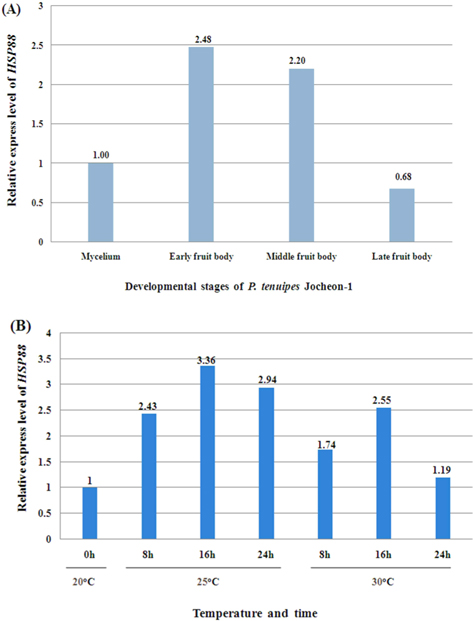
The relative expression level of P. tenuipes Jocheon-1 HSP88 mRNA during the four different developmental stages, mycelium, early fruit body, middle fruit body, and late fruit body, was detected by qPCR. The mycelium stage was used as the control for comparative purposes in normal growth conditions. The highest expression was observed in early fruit body development with expression approximately 2.5 fold higher than that in the mycelium stage and 3.6 fold higher than that in late fruit stage (Fig. 5A).
To ascertain the potential of heat induced expression of P. tenuipes Jocheon-1 HSP88 cDNA, the fruit body of P. tenuipes Jocheon-1 was subjected to heat shock at various temperatures and times. The relative expression level of the P. tenuipes Jocheon-1 HSP88 gene after heat treatment is shown in Figure 5B. During the heat shock treatment of 25℃, the expression level was highest at 16 h, which was 3.36 fold higher than that of the control at the starting point (0 h at 20℃ in the fruit body). During the heat shock treatment at 30°C, the expression level of P. tenuipes Jocheon-1 HSP88 cDNA reached its highest at 16 h, which was 2.55 fold higher than that of the control at the starting point. After 24 h, expression had decreased to levels similar to the control at 24 h.
In this study, the full-length cDNA sequence of HSP88 was cloned from P. tenuipes Jochoen-1. The full-length cDNA of P. tenuipes Jocheon-1 is 2,139 bp and encodes a polypeptide of 713 amino acids. We had previously reported the genomic structure of the HSP88 genes in four entomopathogenic fungi, P. tenuipes Jochoen-1, P. tenuipes, Cordyceps militaris (C. militaris), and Cordyceps pruinosa (C. pruinosa), which are industrialized in Korea (Liu et al., 2012). Among them the three entomopathogenic fungal strains, P. tenuipes Jochoen-1, P. tenuipes, and C. militaris showed identical genomic structure, namely 5 introns and 6 exons with 13, 62, 1,438, 306, and 288 nucleotides encoding 713 amino acid residues. C. pruinosa revealed a different species because it contained 4 introns and 5 exons with the length of 13, 62, 32, 1,744, and 288 nucleotides encoding 713 amino acid residues (Liu et al., 2012). It is very interesting that almost same number of deduced amino acid residues for HSP88 in even different genera or species was identified. Pairwise comparison of P. tenuipes Jocheon-1 HSP88 with those of other fungal strains revealed a wide range of identity ranging from 45-77%. These results demonstrate less identity than in the fruiting body forming insect mushroom species studied previously in 2012 (Liu et al.) This reveals that the HSP genes compared in this study are homologous (even variable in similarity of identity) to the ancestral HSP gene, but there are many mutations in the corresponding sequences that have occurred through long periods of evolutionary time.
The ATPase domains of HSP70, actin, and hexokinase have a common three-dimensional structure (Flaherty et al., 1991). Five sequence motifs named phosphate1, connect1, phosphate2, adenosine, and connect2, which are involved in ATP binding and interdomain coupling (Buchberger et al., 1994), have been identified within these structurally similar domains (Bork et al., 1992). In this study, we found that these motifs are also present in the HSP88 of P. tenuipes Jocheon-1 and other fungal HSP88s, such as the HSP88 of N. crassa (Plesofsky-Vig and Brambl, 1998). Analysis of the amino acid sequence of this gene revealed a considerable identity with the fungal HSP88s of N. haematococca, N. gramincola, and N. crassa. Sequence conservation was highest at the N-terminus and decreased towards the C-terminus. The cDNA encoding P. tenuipes Jocheon-1 HSP88 was expressed as an 88-kDa polypeptide in baculovirus-infected insect Sf9 cells. Future research will continue on the construction of antibodies against HSP88 expressed from the recombinant baculovirus infected with PtHSP88 cDNA and identification of the protein with western blot analysis.
qPCR was performed to detect the relative expression level of P. tenuipes Jocheon-1 HSP88 during the developing stages of mycelium, early fruit body, middle fruit body, and late fruit body, respectively. Results showed that P. tenuipes Jocheon-1 HSP88 has the highest relative expression level during the early fruit body developmental stage. We conducted our experiments at 35℃, but sampling was difficult because of mycerial death at the treated temperature. In organisms that can grow over a broad range of temperatures, the maximum response is usually achieved at about 10℃ above the optimum growth temperature. In organisms that grow over a more restricted range, the maximum response occurs at about 5℃ above the optimum growth temperature. (Lindquist, 1986). The optimum temperature for the growth of P. tenuipes Jocheon-1 during fruit body development is approximately 20℃. Heat shock treatments at 25℃ and 30℃ have been performed and qPCR has been used to analyze the relative expression level of HSPs after heat shock. Results demonstrated that the relative expression level of P. tenuipes Jocheon-1 HSP88 is greatest at 25℃ after 16 h, which is approximately 3.5 times higher than that of control. The relative expression level of P. tenuipes Jocheon-1 HSP88 also increased during heat shock treatment at 30℃ and reached its highest level after 16 h, and then levels reduced similar to those in the control group after 24 h. These results indicate that HSP88 production is probably dependent on both cellular differentiation and heat stress. The results obtained in this research will be useful to further investigate the molecular, biological, physiological, and developmental roles of the HSP88 in entomopathogenic fungi, which are very important species in the industrialization of insect mushrooms.