The genus
Recently, additional specimens of the quadrannulate
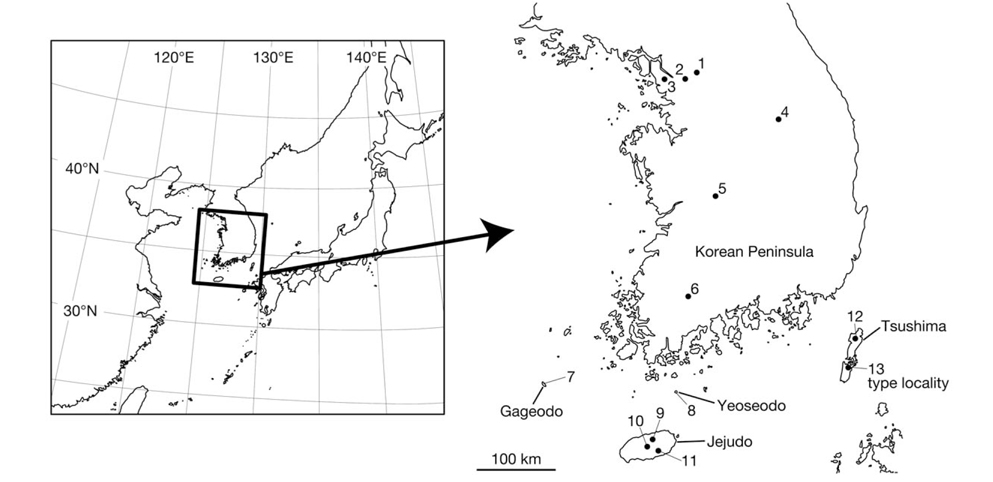
Leeches were collected from various places in Korea (Fig. 1). Altitudes and coordinates for localities were obtained using a Garmin eTrex GPS unit.
For DNA extraction, botryoidal tissue was taken from the posterior part of the body around the caudal sucker of the specimens. All of the specimens were preserved in 70% ethanol. Four measurements were taken: body length (BL) from the anterior margin of the oral sucker to the posterior margin of the caudal sucker; maximum body width (BW); caudal sucker length (CL) from the anterior to the posterior margin of the sucker; caudal sucker width (CW) from the right margin to the left margin of the sucker. Examination, dissection, and drawings of the specimens were accomplished using a stereoscopic microscope equipped with a drawing tube (Leica M125; Leica, Wetzlar, Germany). Specimens used in this study have been deposited in the Zoological Collection of Kyoto University (KUZ).
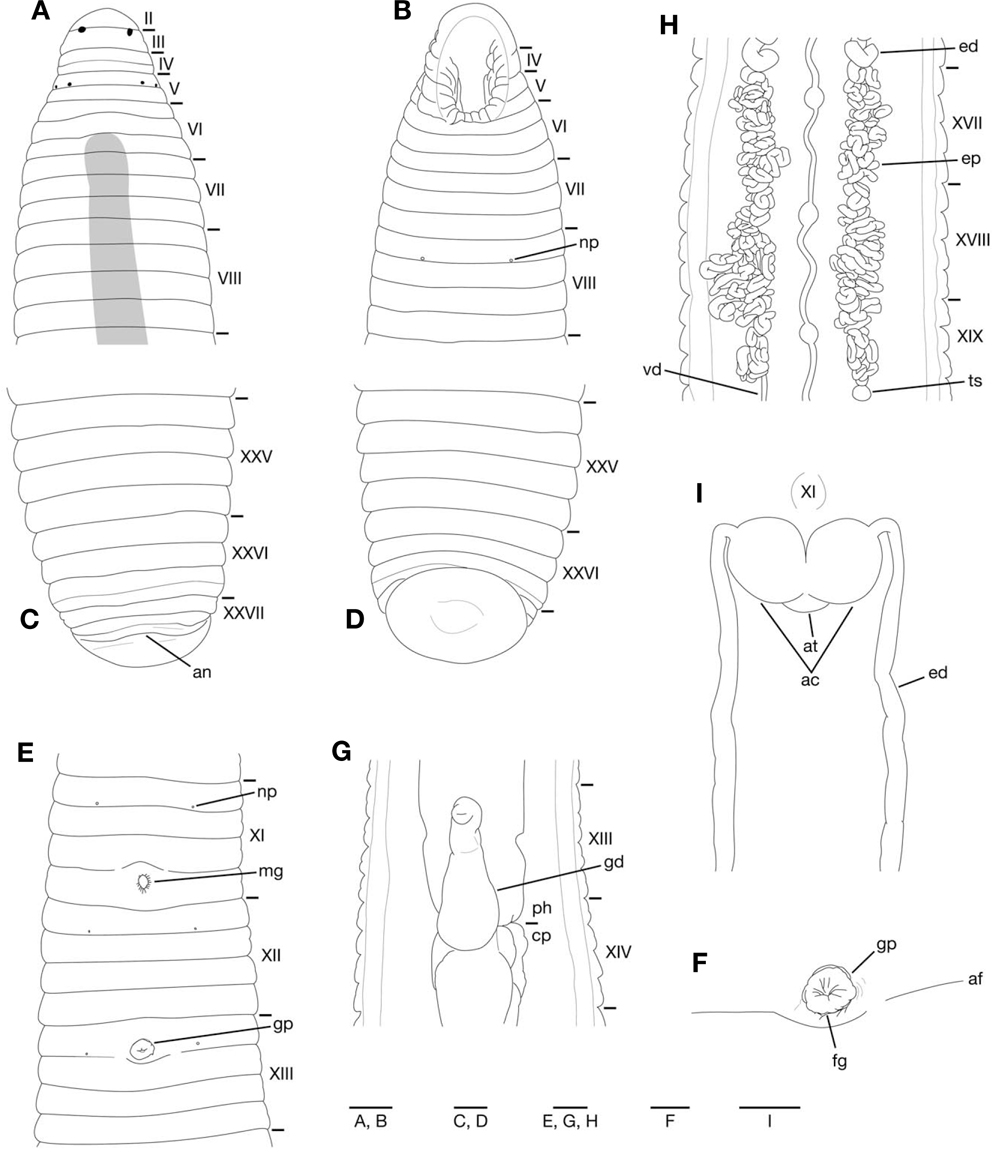
The numbering convention is based on Moore (1927): body somites are denoted by Roman numerals and the annuli in each somite are given alphanumeric designations.
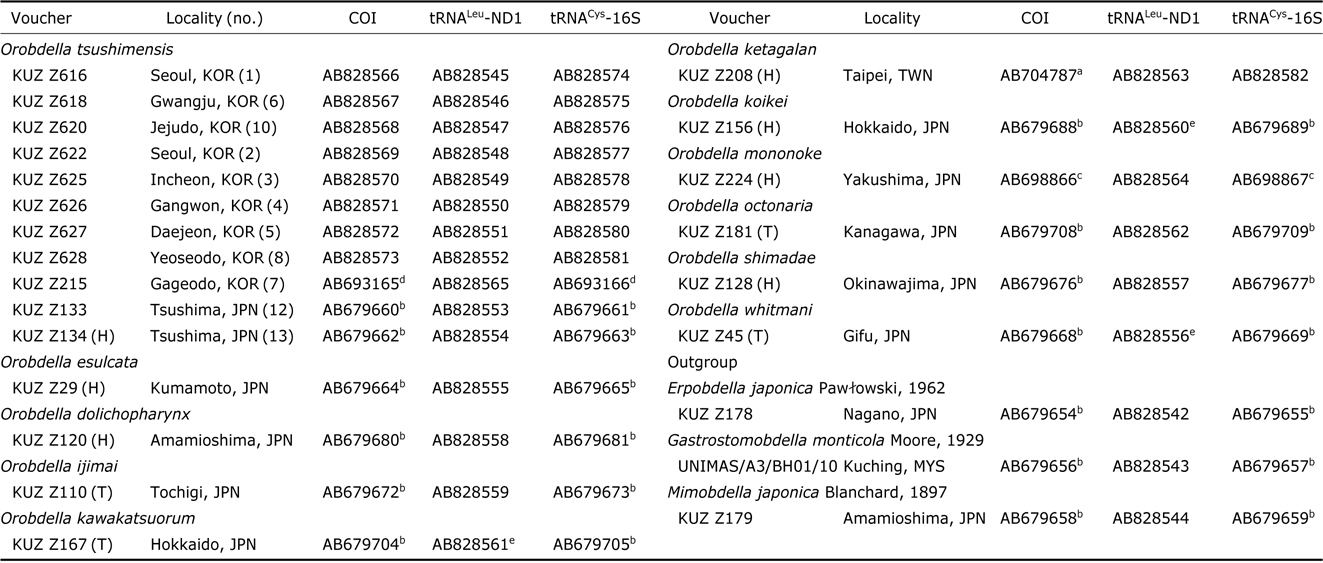
The extraction of genomic DNA and DNA sequencing methods for COI and tRNACys-16S followed Nakano (2012b). For obtaining tRNALeu and ND1 (tRNALeu-ND1) sequence data, the procedure above was modified as follows: the primer sets for tRNALeu-ND1 were LND300 and HND1932 (Light and Siddall, 1999) or HND1932-in (5′-AATGGAGCACGA TTAGTTTC-3′) (in this study); the PCR reaction mixture for tRNALeu-ND1 was heated to 94℃ for 5 min, followed by 35 cycles of 94℃(10 s), 45℃(20 s), and 72℃(39 s) and a final extension at 72℃ for 6 min; the sequencing reaction mixture for tRNALeu-ND1 was incubated at 96℃ for 2 min, followed by 40 cycles of 96℃ (10 s), 50℃ (5 s), and 60℃ (39 s). Obtained sequences have been deposited in GenBank. Sequences used for molecular phylogenetic analyses are listed in Table 1; 16 sequences of COI, 3 those of tRNALeu- ND1 and 15 those of tRNACys-16S, which were published in the previous studies (Nakano, 2012a, 2012b; Nakano and Lai, 2012; Nakano and Seo, 2012; Nakano and Gongalsky, 2014), were obtained from GenBank.
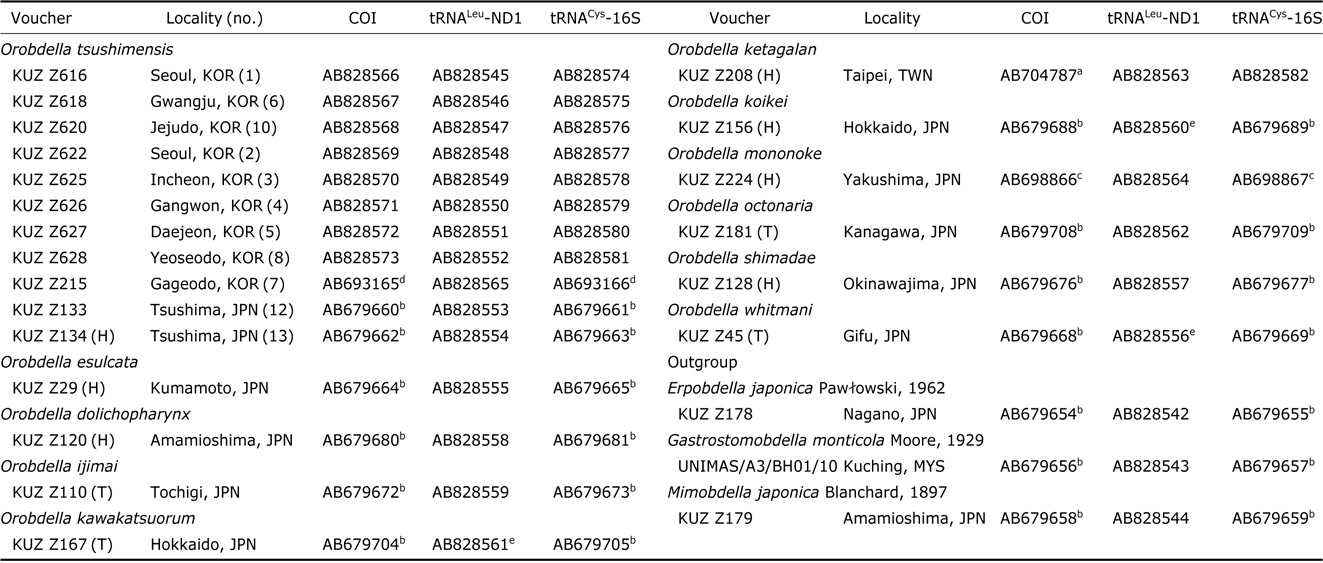
Samples used for the phylogenetic analyses, with the information on voucher and GenBank accession numbers
Sequences of mitochondrial COI and ND1(tRNALeu region was removed from each sequence) were aligned by eye because there were no indels. Mitochondrial tRNACys-16S sequences were aligned using MAFFT L-INS-I (Katoh et al., 2005) and then refined with GBLOCKS (Castresana, 2000). The length of the aligned COI sequences was 1,266 bp, that of ND1 was 579 bp, and that of tRNACys-16S was 818 bp (288 characters were eliminated). The concatenated sequences thus yielded a total of 2,663 bp positions. Pairwise comparisons of Kimura-2 parameter (K2P) distance (Kimura, 1980) of the COI sequences were calculated using MEGA5 (Tamura et al., 2011).
Phylogenetic trees were constructed using maximum likelihood (ML) and Bayesian inference (BI) models. ML phylogenies were calculated using TREEFINDER v October 2008 (Jobb et al., 2004) with the tool package PHYLOGEARS v 2.0 (Tanabe, 2008), and then nonparametric bootstrapping (Felsenstein, 1985) was conducted with 500 replicates. The best-fit models for each partition were selected based on the Akaike Information Criterion (Akaike, 1974) by using KAKUSAN4 (Tanabe, 2011): for the 1st position of COI, TN93 with gamma distribution (+G) and proportion of invariant sites (+I); for the 2nd position of COI, TVM+I; for the 3rd position of COI, HKY85+G; for the 1st position of ND1, GTR+G+I; for the 2nd position of ND1, J1+G; for the 3rd position of ND1, J2+G; and for tRNACys-16S, GTR+G was selected. BI and Bayesian posterior probabilities (BPPs) were estimated using MRBAYES v 3.2 (Ronquist et al., 2012). The best-fit models for each partition were identified under the Bayesian information criterion (Schwarz, 1978) and also by using KAKUSAN4: for the 1st position of COI, GTR+G+I; for the 2nd position of COI, F81+I; for the 3rd position of COI, HKY85+G; for the 1st position of ND1, GTR+G+I; for the 2nd position of ND1, HKY85+G; for the 3rd position of ND1, HKY85+G; and for tRNACys-16S, GTR+G was selected. Two independent runs of four Markov chains were conducted for 4 million generations, and the tree was sampled every 100 generations. The parameter estimates and convergence were checked using TRACER v 1.5 (Rambaut and Drummond, 2009), and based on these results, the first 10,001 trees were discarded.
Nodes with bootstrap (BS) values higher than 70% were considered as sufficiently resolved (Hillis and Bull, 1993). Nodes with BPPs higher than 95% were considered statistically significant (Leaché and Reeder, 2002).
Order Arhynchobdellida Blanchard, 1894 Suborder Erpobdelliformes Sawyer, 1986 1*Family Gastrostomobdellidae Richardson, 1971 2*Genus Orobdella Oka, 1895
3*Orobdella tsushimensis Nakano, 2011 (Figs. 2, 3)
Material examined (see Fig. 1 for the locality number). Korea: 3 specimens, Seoul: Nowon-gu, Mt. Suraksan, alt. 118 m, 37° 41′08′′N, 127° 04′07′′E (locality no. 1), 4 Jul 2012, Seo HY, KUZ Z615, Z616, dissected, and KUZ Z617; 2 specimens, Jongno-gu, Mt. Bukhansan, alt. 447 m, 37° 37′ 28′′N, 126° 57′41′′E (locality no. 2), 6 Jul 2012, Seo HY, KUZ Z621 and Z622, dissected; 2 specimens, Jeollanam-do: Sinan-gun, Gageodo Island, alt. 429 m, 34° 04′49′′N, 125° 96′27′′E, (locality no. 7), 10 Jul 2012, Seo HY, KUZ Z623 and Z624; 1 specimen, Wando-gun, Yeoseodo Island, alt. 46 m, 33° 59′03′′N, 126° 55′19′′E (locality no. 8), 20 Jul 2012, Seo HY, KUZ Z628, dissected; 1 specimen, Jejudo Island: Jeju-si, alt. 384 m, 33° 26′47′′N, 126° 33′49′′E (locality no. 9), 17 Jun 2012, Seo HY, KUZ Z614; 1 specimen, Seogwipo-si, alt. 183 m, 33° 18′05′′N, 126° 37′51′′E (locality no. 11), 27 Sep 2012, Seo HY, KUZ Z629; 1 specimen, Jeju-si, Mt. Hanlasan, alt. 1,601 m, 33° 21′56′′N, 126° 30′48′′E (locality no. 10), 2 Jul 2012, Kang SG, KUZ Z620, dissected; 1 specimen, Gwangju Metropolitan City: Dong-gu, Mt. Mudeungsan, alt. 533 m, 35° 07′11′′N, 126° 58′56′′E (locality no. 6), 1 Jul 2012, Seo HY, KUZ Z618, dissected; 1 specimen, Dong-gu, Mt. Mudeungsan, alt. 375 m, 35° 07′21′′N, 126° 58′34′′E (locality no. 6), 24 Jun 2012, Seo HY, KUZ Z619; 1 specimen, Incheon Metropolitan City: Gyeyang-gu, Mt. Gyeyangsan, alt. 63 m, 37° 33′33′′N, 126° 43′30′′E (locality no. 3), 11 Oct 2012, Seo HY, KUZ Z625, dissected; 1 specimen, Gangwon-do: Wonju-si, Mt. Chiaksan, alt. 790 m, 37° 18′07′′N, 128° 03′39′′E (locality no. 4), 5 Oct 2012, Seo HY, KUZ Z626, dissected; 1 specimen, Daejeon Metropolitan City: Seo-gu, Mt. Changtaesan, alt. 245 m, 36° 11′38′′N, 127° 20′19′′E (locality no. 5), 5 Oct 2012, Seo HY, KUZ Z627, dissected.
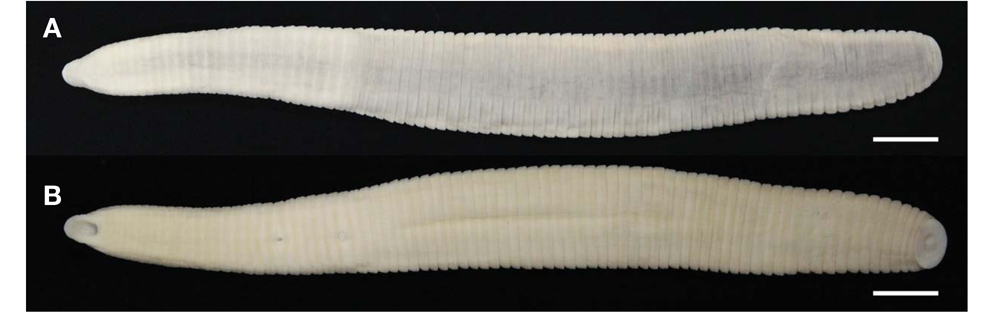
Description. Maximum BL 105.4 mm (KUZ Z622), BW 11.9mm(KUZ Z628), minimum BL 33.2 (KUZ Z626), BW 4.3mm(KUZ Z619). Caudal sucker ventral, ellipsoid, maximum CL 3.9 mm, CW 6.6 mm (KUZ Z622), minimum CL 1.6mm(KUZ Z624), CW 2.0mm(KUZ Z626).
Somites VIII-XXV quadrannulate, somite XXVI triannulate (Fig. 3A-E). Anus behind somite XXVII with no postanal annulus (Fig. 3C). Clitellum in XI b5 to XIII a2. Eyes often in 3 pairs, first pair dorsally in posterior margin of II, second and third pairs dorsolaterally in posterior margin of V (a1+a2) (Fig. 3A). Nephridiopores in 17 pairs in VIII-XXIV (Fig. 3B, E).
Pharynx reaching to XIII b5/b6-XIV b6 (Fig. 3G); each pharyngeal ridges reaching to crop straightly. Crop tubular, reaching to XIX b5/b6-XX a1/a2. Gastropore conspicuous in XIII a1/a2 (Fig. 3E, F). Gastroporal duct developed, bulbous, or bottle-shaped, slightly winding at junction with gastropore; joining with crop in XIII b6-XIV b6 (Fig. 3G).
Male gonopore in XI b6 (slightly anterior to middle of annulus), female gonopore inconspicuous in XIII a1/a2, behind gastropore, gonopores separated by 1/2+5 annuli (Fig. 3E, F). Testisacs multiple, in XVIII b5-XX b6 to XXIV a1- XXVb6(Fig. 3H). Paired epididymides in XV b5-XVIII a1/a2 to XVIII b5-XX a1/a2 (Fig. 3H). Ejaculatory ducts in XI b5 to XV b5-XVIII a1/a2; nearly straight in position anterior to ovisacs (Fig. 3I); coiled in position posterior to ovisacs (Fig. 3H). Pair of muscular atrial cornua, ovate in XI b5 and b6 (Fig. 3I) or fusiform in XI b6. Atrium short, muscular, globular, in XI b5 and b6. One pair of ovisacs, thin-walled, globular, in XIII a2 and b5. Oviducts thin-walled, both oviducts converging into common oviduct in XIII a2.
Distribution. Known from mountainous regions of the Korean Peninsula and its peripheral islands including Tsushima Island, Japan (Nakano, 2011a; Nakano and Seo, 2012).
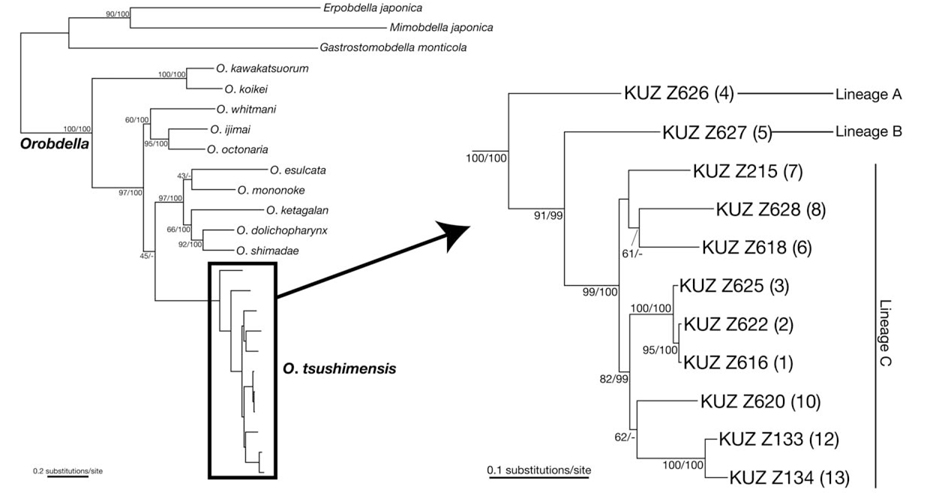
Phylogenetic relationships (see Fig. 1 for the locality number). The ML tree with ln
Monophyly of
Genetic divergence. The COI K2P distance within
Remarks. The present specimens newly collected from various places in Korea clearly belong to
The molecular phylogenies obtained here showed that the specimens that were morphologically identified as
The present molecular phylogenies showed that the specimen from Mt. Chiaksan (Lineage A; locality no. 4) first diverged from the others. Then, the specimen from Mt. Changtaesan (Lineage B; no. 5) diverged from the other leeches. It is noteworthy that the specimen from Jejudo Island (no. 10) and two specimens from Tsushima Island (nos. 12, 13) are monophyletic, which was statistically supported, with the three specimens from locations around Seoul (nos. 1-3), not with the specimens from adjacent localities (i.e., nos. 6-8). This geographic pattern of the phylogenetic relationships suggests a complex biogeographical history of