For livestock farming, antimicrobial agents are abundantly used not only for medical treatment and prophylaxis of bacterial infections but also in the animals’ food to promote growth, which has resulted in an increase in antimicrobial-resistant bacterial strains (Aarestrup, 2005; Sapkota et al., 2008). The critical factor in the development of antimicrobial resistance is the ability of the bacteria to acquire and disseminate exogenous genes via mobile genetic elements, such as plasmids, transposons, DNA insertion elements, and genomic islands (Rowe-Magnus et al., 2002). Horizontal gene transfer is one of the most common mechanisms, by which antimicrobial resistance traits are transferred from one organism to another (Martinez, 2009). Since the discovery of integrons, a unique mechanism for the dissemination of antibiotic resistance genes (Stokes and Hall, 1989), the increase in bacterial strains resistant to several antimicrobial agents has raised public health concerns about treatment options, heath care costs, and foodborne illness (Hammerum and Heuer, 2009). Food contamination of antimicrobial-resistant bacteria could be a major public health threat, due to the possibility that the responsible genes, which are carried on mobile genetic elements, may be transferred to other bacteria that confer high risks to human health (Rowe-Magnus et al., 2002).
In Korea, numerous studies have focused on antimicrobial resistance patterns and genes in
In this study,
>
E. coli isolation and identification
In total, 131 environmental strains of
>
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing against 22 antimicrobials was performed on 131
>
PCR amplification of antimicrobial resistance and virulence genes
The presence of genes associated with tetracycline (
>
Isolation of E. coli strains from surface seawater in Gomso Bay
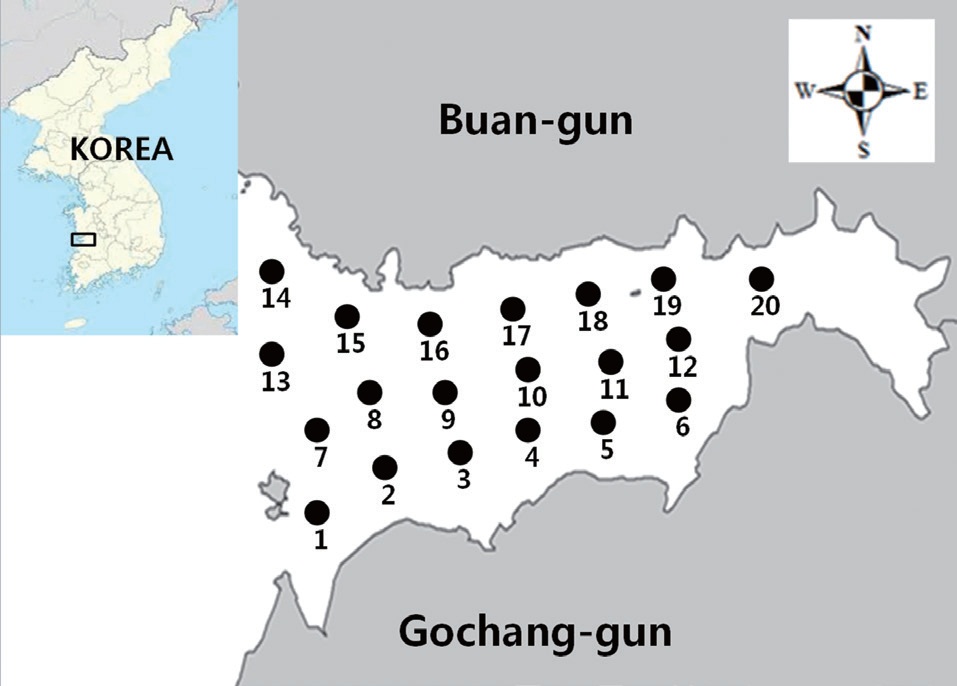
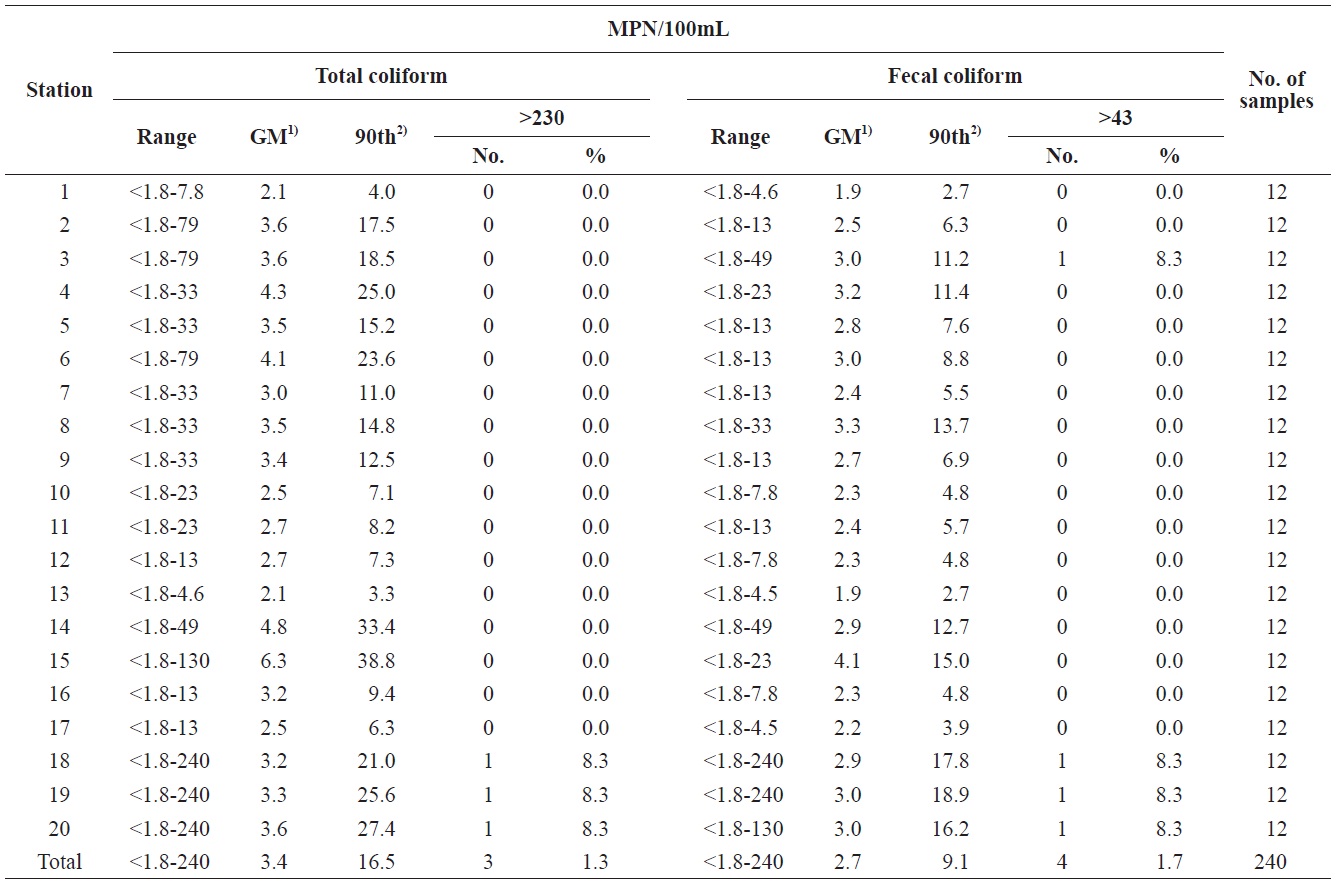
A sanitary survey in an important shellfish-growing area of Gomso Bay, west coast of Korea, was conducted to evaluate bay conditions and compliance with bacteriological criteria for shellfish production. Seawater samples were collected 12 times from May to December 2012 from 20 sampling stations established in the survey area (Fig. 1). The values determined by the MPN-method for both the total coliform bacteria and fecal coliform bacteria in the 240 seawater samples ranged from less than 1.8 to 240 MPN/100 mL, respectively (Table 2). The geometric means and estimated 90th percentile values for total coliform bacteria ranged from 2.1 to 6.3 MPN/100 mL, and from 3.3 to 38.8 MPN/100 mL, respectively, whereas those for fecal coliform bacteria ranged from 1.9 to 4.1 MPN/100 mL,
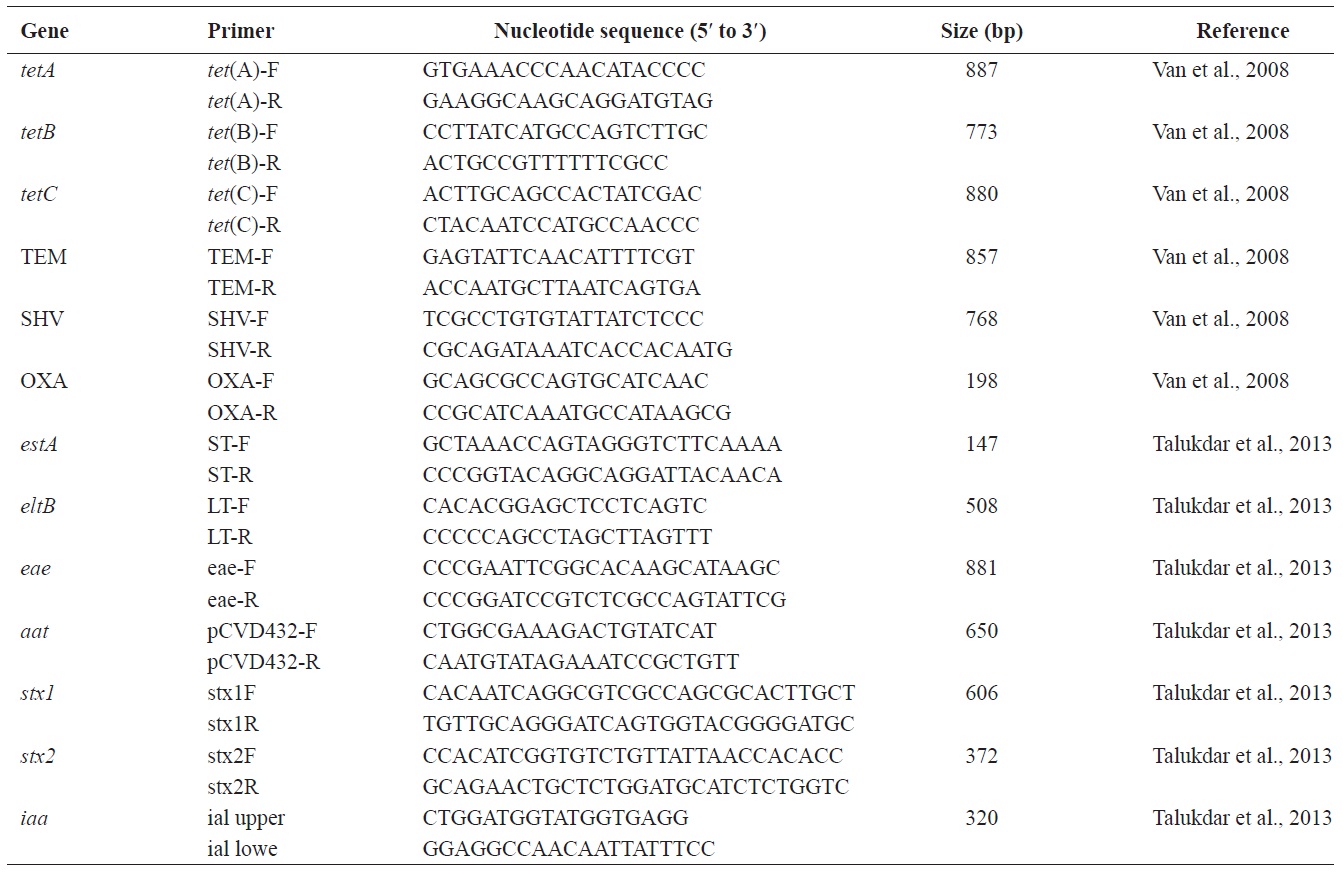
[Table 1.] Oligonucleotide primers used in this study
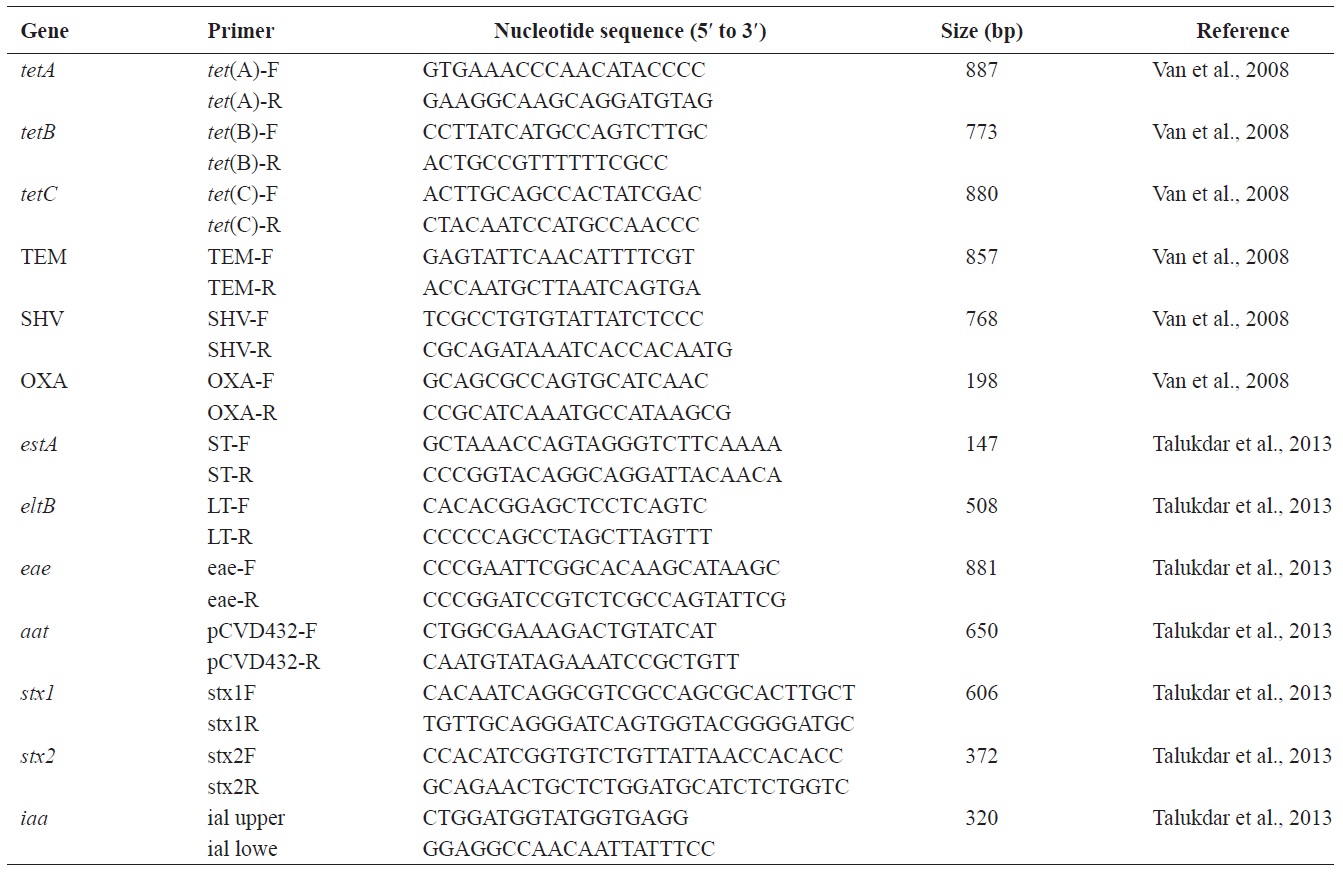
Oligonucleotide primers used in this study
and from 2.7 to 18.9 MPN/100 mL, respectively. Notably, the levels of total coliform and fecal coliform bacteria from stations near the wastewater discharge sites (stations 3, 14, 18, 19, and 20) were slightly higher than those from other sites. The bacteria levels after rainfall were also higher, than those in the dry season. Based on these data, the bacteriological quality of seawater in Gomso Bay met the criteria recommended by the National Shellfish Sanitation Program (Food and Drug Administration, 2005) and the Korea Shellfish Sanitation Program for shellfish production (Ministry of Marine Affairs and Fisheries, 2006). One strain from each seawater sample was isolated to further analyze antimicrobial resistance and virulence genes in isolated
>
Antimicrobial susceptibility of E. coli isolates
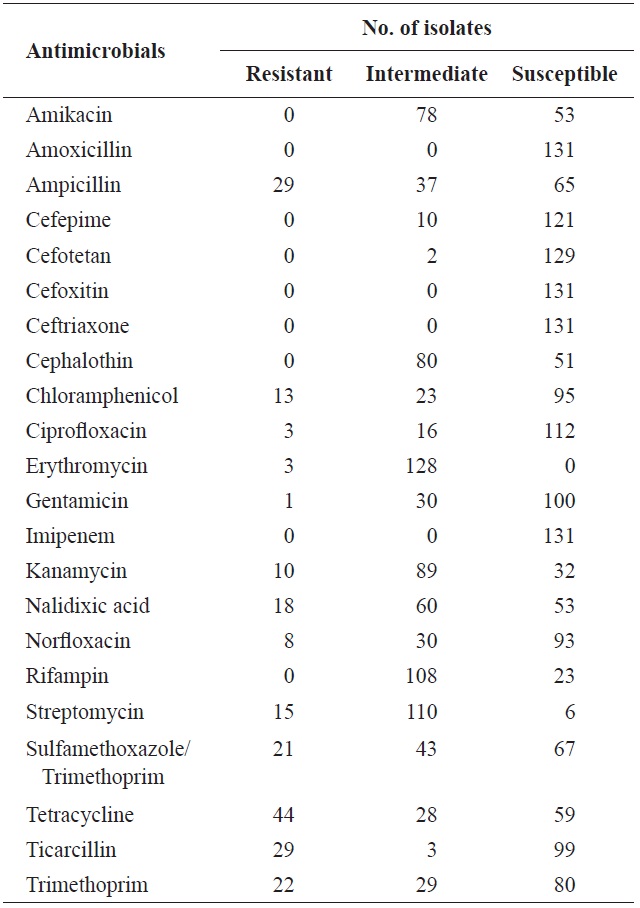
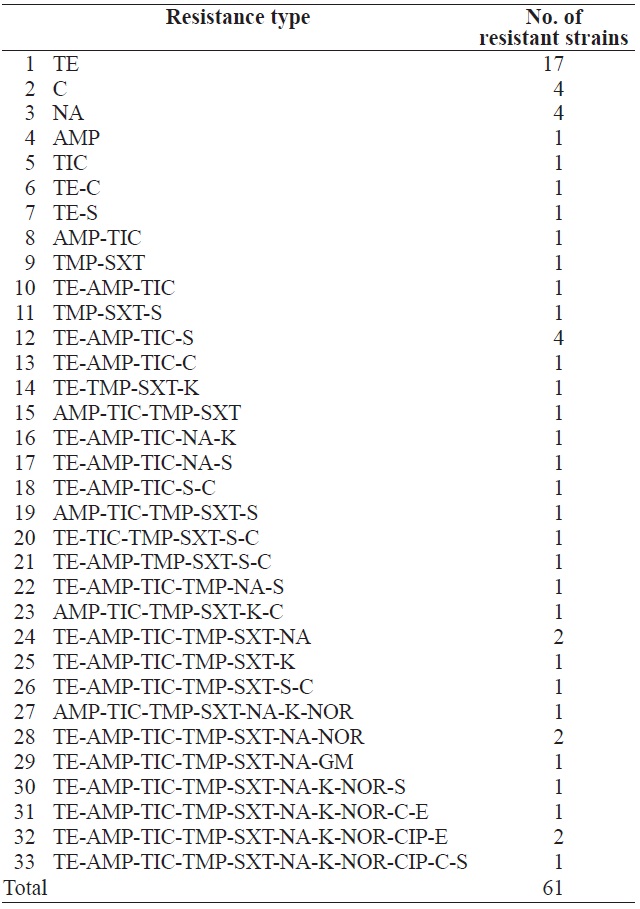
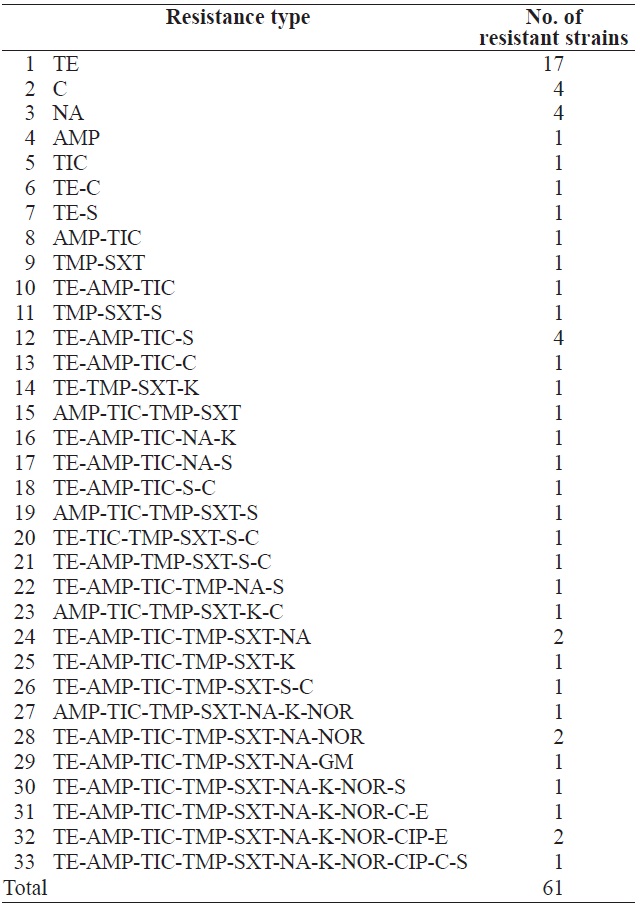
In total, 131 isolates were tested for resistance against 22 antimicrobial agents (Table 3). Resistance to tetracycline (33.6%), ampicillin (22.1%), ticarcillin (22.1%), trimethoprim (16.8%), sulfamethoxazole/trimethoprim (16.0%), nalidixic acid (13.7%), streptomycin (11.5%), chloramphenicol (9.9%), kanamycin (7.6%), norfloxacin (6.1%), ciprofloxacin (2.3%), erythromycin (2.3%), and gentamicin (0.8%) was observed. These results are in agreement with several previous studies (You et al., 2006; Van et al., 2008; Koo and Woo, 2012; Ryu et al., 2012b). However, no isolate showed resistance to amikacin, amoxicillin, cefepime, cefotetan, cefoxitin, ceftriaxone,
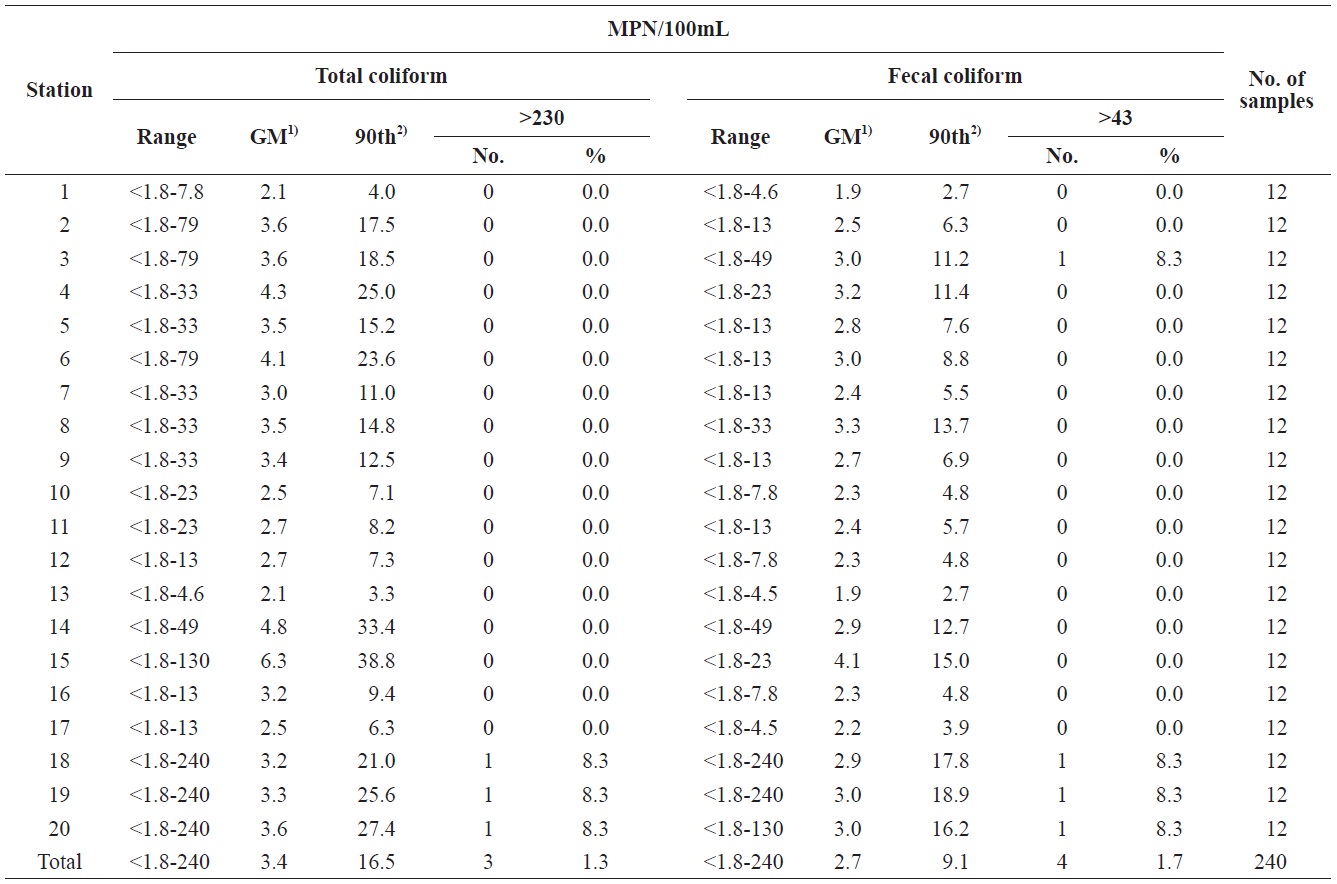
Summary of bacteriological examination results of each sampling stations in Gomso Bay, from May to December 2012
cephalothin, imipenem, or rifampin. Table 4 shows the resistance patterns of these isolates. Among the 131
>
Characterization of antimicrobial resistance genes in E. coli isolates
Ampicillin-resistant isolates (
Among the 44 tetracycline-resistant
>
Detection of virulence genes by PCR
All 131 isolates were also examined for the presence of seven virulence genes (
[Table 3.] Antimicrobial resistance of Escherichia coli isolated from surface seawater in Gomso Bay
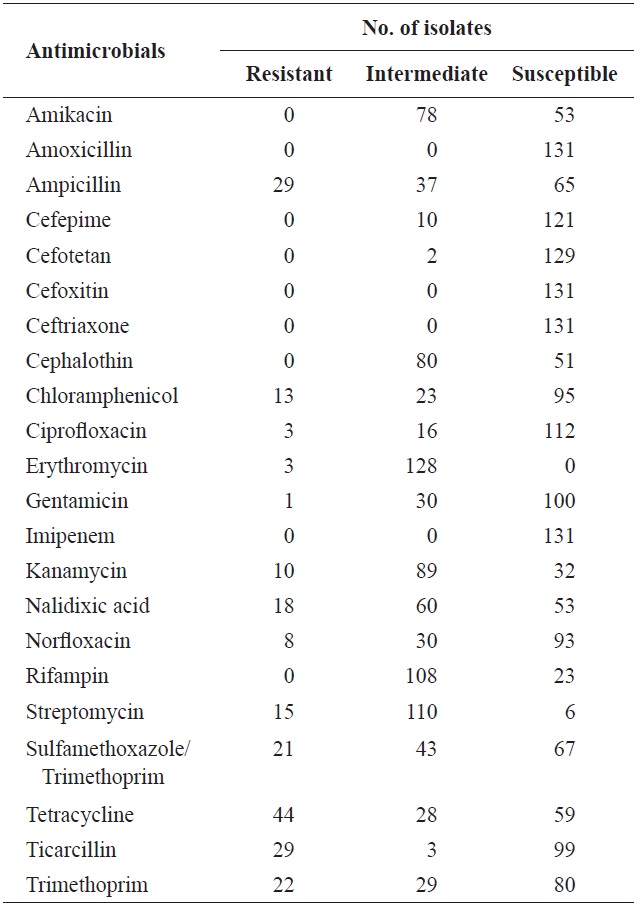
Antimicrobial resistance of Escherichia coli isolated from surface seawater in Gomso Bay
2.3% (
Our current study focused primarily on the prevalence of
Antimicrobial resistance patterns of Escherichia coli isolated from surface seawater in Gomso Bay
a source of shellfish and other aquaculture organisms. Waste from agriculture, household and stock breeding of numerous small rivers that flow into Gomso Bay are thought to be the current source of contaminants in the area.
Virulent or antimicrobial-resistant bacteria in drainage water can contaminate shellfish and fisheries in the bay and potentially be hazardous for human health. Many antibiotics are used for therapeutic treatment and prophylactic purposes in animals and humans; consequently, increase in antimicrobial resistance due to excess use and misuse of antimicrobials is a public health concern (World Health Organization, 2001). Therefore, antimicrobial usage in animal feed should be tightly regulated to decrease the prevalence of antimicrobial-resistant bacteria.
In this study, 131 environmental
Ampicillin resistance in gram-negative bacteria is primarily mediated by β-lactamases, which hydrolyze the β-lactam ring and thereby inactivate antibiotics (Massova and Mobashery, 1998). In many genera of gram-negative bacteria, the most predominant β-lactamases are TEM-, SHV-, CTX-, and OXA-type enzymes (Bradford, 2001). TEM-1, a plasmid-mediated β-lactamase, is the most common β-lactamase in gram-negative bacteria, and accounts for up to 90% of the ampicillin resistance in
In conclusion, continuous surveillance of antimicrobial-resistant bacteria in Gomso Bay is necessary to ensure the safety of fisheries and shellfish production.