The interactions of metal cations with DNA, as a part of a [M·DNA] complex, have been studied extensively using IR,1X-ray,2 and other techniques.3 Metal cations can both stabilize and destabilize DNA;4 they prefer to interact with the phosphate anions in the backbone of DNA by nonspecific electrostatic attraction, which stabilizes the DNA helix structure. 1,5 However, several divalent metal cations destabilize DNA and decrease the mean transition temperature (Tm) of DNA. 4 In particular, Cu cations substantially decrease the Tm of DNA.4 Cu cations are regarded to be effective in destroying the native structure of DNA. In contrast to Cu cations, Mg cations increase the Tm of DNA and stabilize DNA.
The structures of [M·(nucleoside monophosphate)] complexes have been investigated to evaluate the effect of metal cations on the structure of the [M·DNA] complex.6-11 Metal cations prefer the phosphate anions; however, several complexperimental results showed that the base moieties had also been regarded as the preferred binding sites in the formation of [M·(nucleoside monophosphate)] complexes. As a member of [M·(nucleoside monophosphate)] complexes, [M·dCMP] (dCMP = deoxycytidine monophosphate) complexes have been extensively studied.7-11 A local pentacoordinated tetragonal pyramid geometry with a coordination of one N atom (cytosine) and four O atoms has been observed by the powder EPR spectrum of [Cu·CMP] complex.7 The metal binding sites of the [M·H·dCMP]1+ (M = Mg2+ , Cu2+ ) complex has been observed in the aqueous solution; 10 the metal cation was mainly located at the N3 (cytosine) atom, as determined by the acidity constant analysis using potentiometric pH titrations. However, the coordination chemistry between Cu cation and nucleic acid building block such as nucleotides, is rather scarcely documented in gas-phase.
In this study, we have focused our attention on the formation and fragmentation pattern of gas-phase [dCMP]1- and [M·dCMP·dCMP-H]1- (M = Mg2 +, Cu2+) complexes using ESI-MS and tandem mass spectrometry (MS/MS) methods. The [dCMP]1- and [M·dCMP·dCMP-H]1- complexperimental exes were formed in the solution and electrosprayed on the quadrupole ion guide using nitrogen gas. Intact gas-phase [dCMP]1- and [M·dCMP·dCMP-H]1- complex ions were expected in the ESI-MS spectra. 12-14 An
Intact gas-phase [dCMP]1- or [M·dCMP·dCMP-H]1- (M = Mg2+, Cu2+) ion was formed by the ESI-MS method. The experimental MS and MS/MS data for the fragmentation pattern analysis were obtained using a Thermo Finnigan LTQ mass spectrometer (Thermo Electron Corporation, San Jose, CA, USA). The LTQ mass spectrometer conditions have reported in the previous study.15
The reagents used are as follows: dCMP (2’-deoxycytidine 5’-monophosphate 98%, Sigma-Aldrich, Korea), CuCl2 (99.999%, Sigma-Aldrich, Korea), MgCl2 (99.99%, Sigma-Aldrich, Korea), H2O (HPLC grade, Merck), and D2O (99.9 atom%, Sigma-Aldrich, Korea). dCMP was dissolved in H2O (or D2O) to prepare a 4×10-5 M solution. CuCl2 was dissolved in H2O (or D2O) to prepare a 4×10-4 M solution. The [dCMP + metal] solutions were mixed prior to the mass spectral analyses.
The
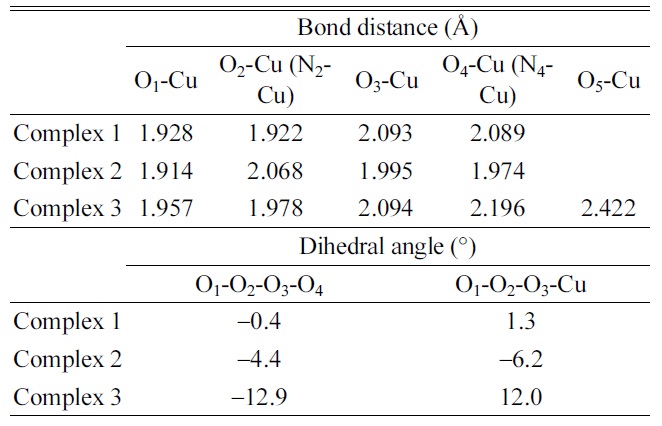
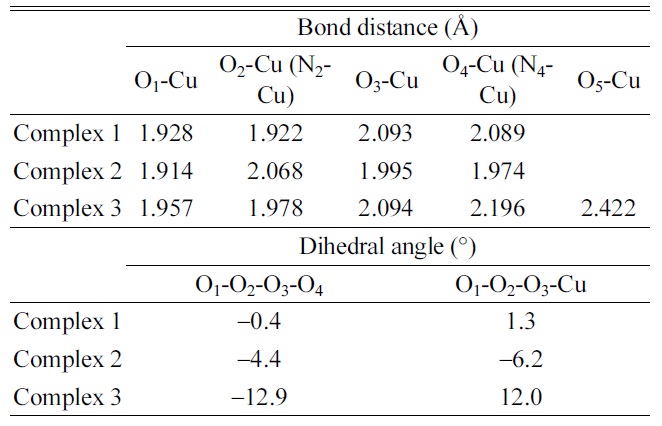
The structures of [dCMP]1- and three [Cu·dCMP·dCMPH]1- complexes are shown in Fig. 1. A copper ion is located in the center of two dCMP molecules because of the basicity of the corresponding phosphate groups. Complexes 1 and 2 show a square planar geometry, where the Cu cation is tetra-coordinated to the four O atoms in Complex 1, or two O and two N atoms in Complex 2. Complex 3 showed a tetragonal pyramid geometry, where the Cu cation is penta-coordinated to the five O atoms. The N3 atom of cytosine in dCMP is indicated as a part of the [Cu...dCMP] coordination in Fig. 1c.
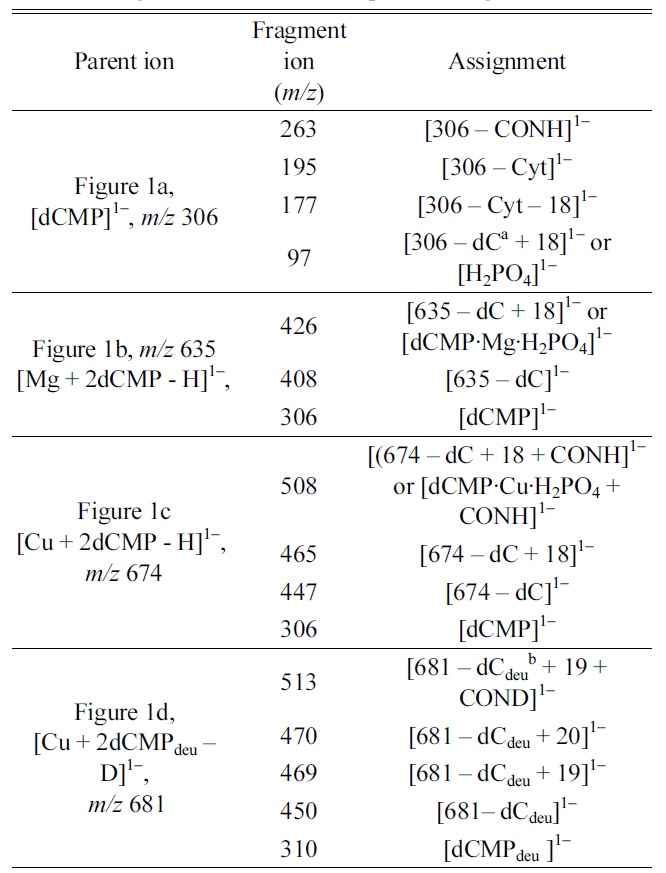
Four MS/MS spectra are shown in Fig. 2. In the MS/MS spectrum of [dCMP]1- ion (Fig. 2a), the fragmentions at
The MS/MS spectrum of the [Mg·dCMP·dCMP-H]1− complex (
may have originated from a similar dissociation process. Based on the observation in Fig. 2b, Complex 1 geometry (dCMP...Mg...dCMP) should be regarded as a stable geometry in the [Mg·dCMP·dCMP-H]1− complex.18
Fig. 2c shows the MS/MS spectrum of the parent [Cu·dCMP·dCMP-H]1− (
Other main MS/MS fragmentions of the parent [Cu·dCMP·dCMP-H]1− (
In order to know the stability of the [Cu·dCMP·dCMP- H]1− complex ion, we tried to optimize the geometries of Complexes 1-3 by the
[Table 1.] Fragment ions in MS/MS spectra of Figs. 2a?2d
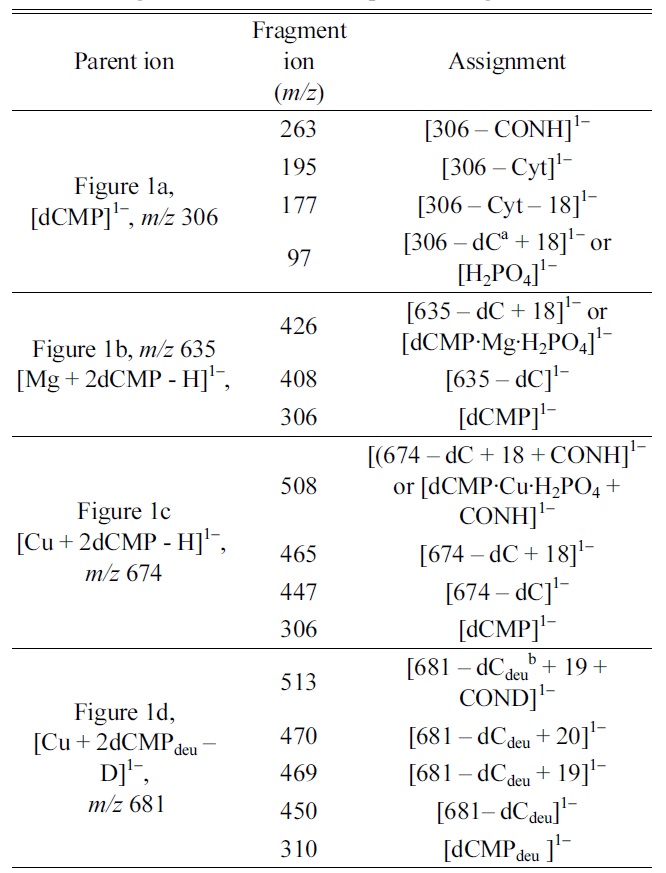
Fragment ions in MS/MS spectra of Figs. 2a?2d
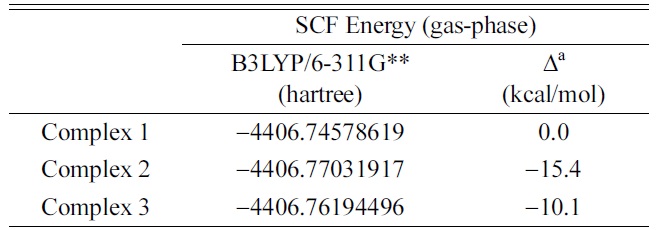
SCF energies of three optimized [Cu·dCMP·dCMP ? H]1? complex ions in B3LYP/6-311G** calculations
[Table 3.] Atomic charge distributions of optimized [dCMP]1? ion
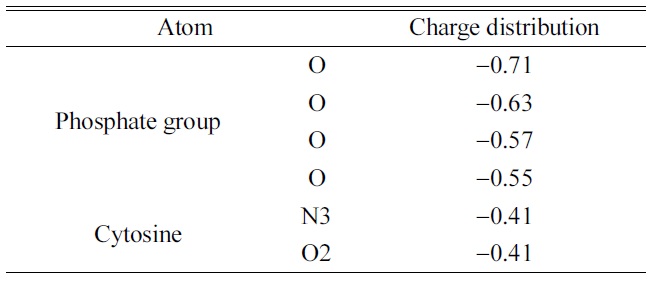
Atomic charge distributions of optimized [dCMP]1? ion
Optimized geometric parameters of [Cu·dCMP·dCMPH1? complex ions in B3LYP/6-311G** calculations
The planar geometries (Cu-O1O2O3O4 for Complex 1, Cu-O1N2O3N4 for Complex 2, and Cu-O1O2O3O4 for Complex 3) were observed for the small dihedral angles in the geometry optimization of the [Cu·dCMP·dCMP− H]1− complex. The optimized parameters such as bond distances and dihedral angles are listed in Table 4. The dihedral angles, O1-O2-O3-O4 (−0.4o) for Complex 1 and O1-N2-O3-N4 (−4.4o) for Complex 2, were almost zero. The structure of complex 3 (Cu- O1O2O3O4O5) was optimized to a tetragonal pyramidal geometry (Fig. 1d). A fifth ligand (the cytosine O2 atom, O5) was added to the four-coordination (Cu-O1O2O3O4) geometry in the tetragonal pyramidal geometry. The carbonyl oxygen atom (O5) of cytosine was located at the apex of the optimized tetragonal pyramidal geometry. A small deviation from the planar structure of Complex 3 was observed in the dihedral angles, O1-O2-O3-O4 (− 12.9o) and O1-O2-O3-Cu (12.0o) (Table 4).
The Cu-binding site in the [Cu·dCMP·dCMP-H]1− complex was investigated. In the MS/MS spectrum of the [Cu·dCMP·dCMP-H]1− complex, the [dCMP·Cu·H2PO4 + CONH]1− fragmention was observed as the main fragmention. Based on the direct interaction between the Cu cation and CONH in the [dCMP·Cu·H2PO4 + CONH]1− fragmention, we proposed that the Cu cation is bound simultaneously to the cytosine (N3 or O2) and a phosphate group. The simultaneous coordination of Cu cation to the phosphate site and cytosine moiety (Complex 2 or 3) was supported by the SCF energy calculations of the geometry optimized [Cu·dCMP·dCMPH]1− complexes.
![Structures of [dCMP]1? and [Cu·dCMP·dCMP-H]1? complexes.](http://oak.go.kr/repository/journal/12929/E1MPSV_2013_v4n4_67_f001.jpg)
![ESI-MS/MS spectra of (a) [dCMP]1? , (b) [Mg + 2dCMPH] 1? , (c) [Cu + 2dCMP - H]1? , and (d) [Cu + 2dCMPdeu -D]1? parent ion.](http://oak.go.kr/repository/journal/12929/E1MPSV_2013_v4n4_67_f002.jpg)
![SCF energies of three optimized [Cu·dCMP·dCMP ? H]1? complex ions in B3LYP/6-311G** calculations](http://oak.go.kr/repository/journal/12929/E1MPSV_2013_v4n4_67_T002.jpg)
![Atomic charge distributions of optimized [dCMP]1? ion](http://oak.go.kr/repository/journal/12929/E1MPSV_2013_v4n4_67_T003.jpg)