The genus
Since Ehrenberg (1831, 1832) described six
shape, and internal organization (Bourelly 1970). However, it is difficult to delimit
Hill (1991) revised the broad generic definition of the genus
In this study, we report unrecorded Korean
>
Algal cultures and microscopy
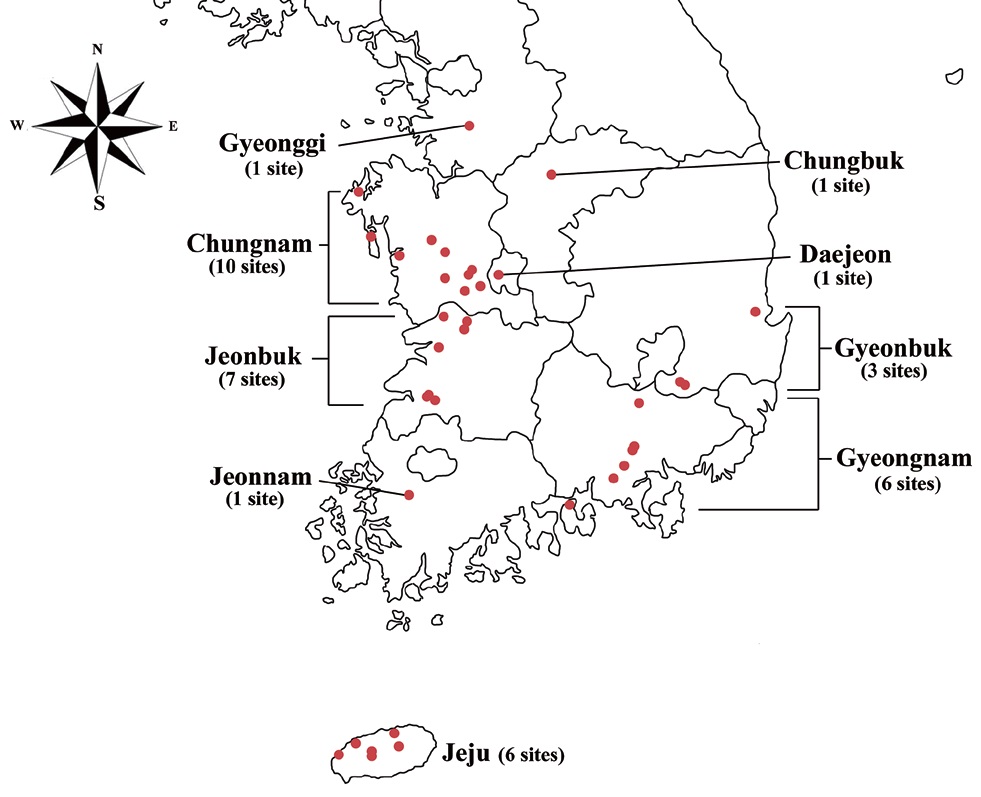
Specimens were collected from freshwater habitats in Korea (Fig. 1). Live cells were isolated by Pasteur capillary pipette and were brought into a unialgal culture. The cells were cultivated in f/2 medium (Guillard and Ryther 1962, Guillard 1975) with soil extract. The clonal cultures were maintained under a light : dark regime of 14 : 10 at 20-22℃ using cool-white fluorescence lamps with illuminations of 30 μmol protons m-2 s-1. The morphology was examined by differential interference contrast with a 100X oil immersion lens (Carl Zeiss Co., Gottingen, Germany). Calibration of magnification was done with grated micrometer. The shape and length of cells, length of the fullow-gullet system, color and number of chloroplasts, presence / absence and number of pyrenoids were examined. Cellular dimensions were determined by measuring 20-25 cells of each taxon from photographic images. Light micrographs were taken with an Axio CamHRc (Carl Zeiss Co.) photomicrographic system attached to the microscope.
>
DNA isolation, polymerase chain reaction (PCR), and sequencing
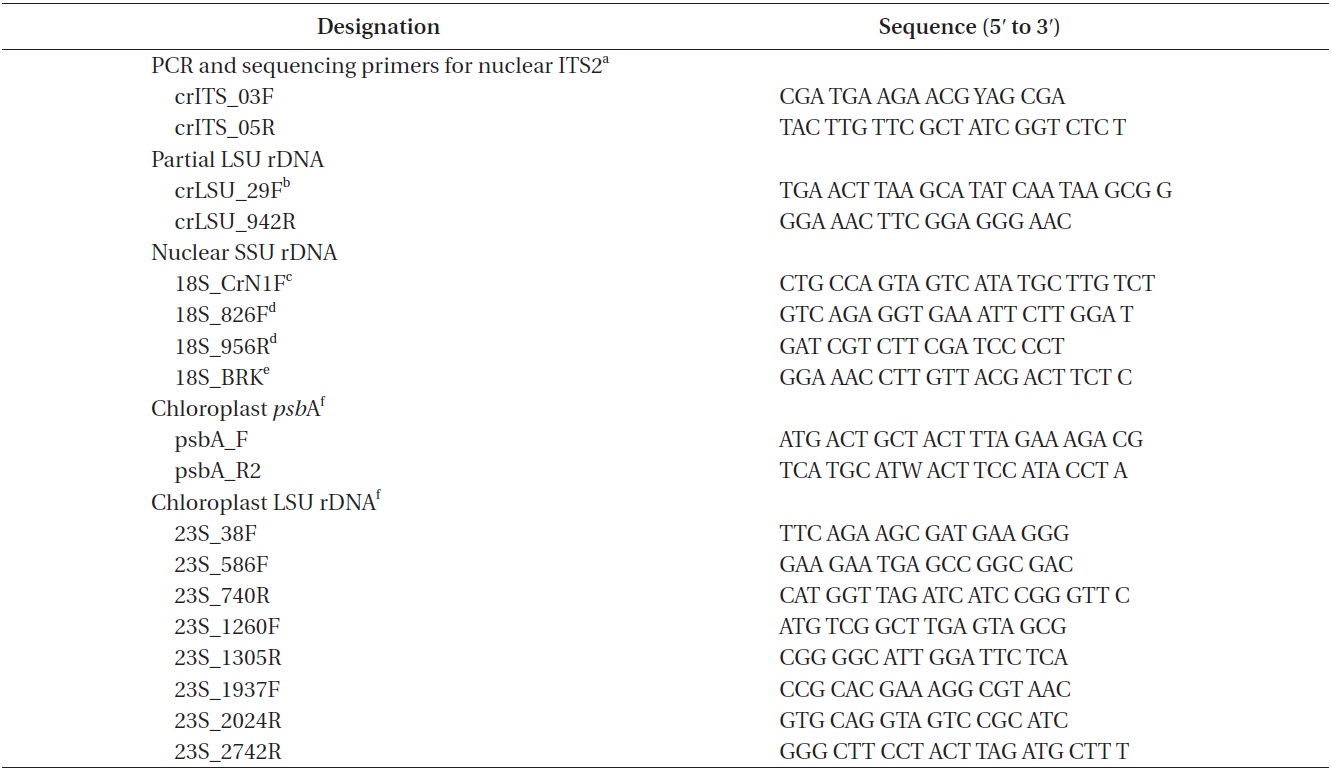
Approximately 10 mL of cultures in exponential growth were harvested by centrifugation (4,500 ×g, model 5415; Eppendorf, Hamburg, Germany) for 1 min at room temperature and washed three times with sterilized distilled water. Total genomic DNA was extracted from the pellet using the Dokdo-Prep Blood Genomic DNA Purification Kit (Elpis-Biotech Inc., Daejeon, Korea) following the manufacturer’s blood sample protocol. PCR was performed using specific primers for nuclear ITS2, nuclear SSU rDNA, chloroplast
[Table 1.] PCR and sequencing primers
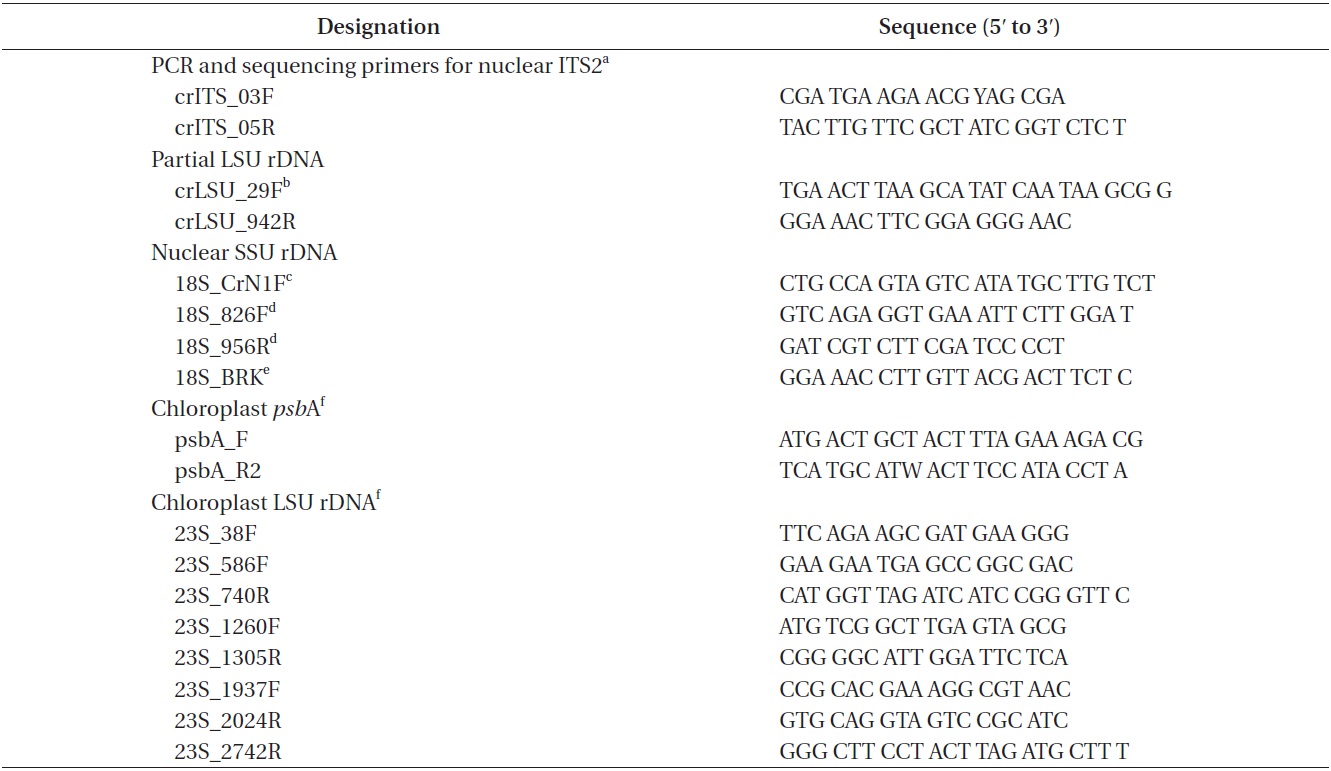
PCR and sequencing primers
The alignment for each gene sequence was aligned by the eye, and was edited using the Genetic Data Environment (GDE 2.4) program (Smith et al. 1994). Unalignable nucleotides were excluded from phylogenetic analyses.
Nuclear ITS2 is likely a suitable marker to identify species according to its degree of conservation (Hoef-Emden 2007), and nuclear ITS2 as well as partial LSU rDNA sequences were used to examine groups of genetically identical strains and to identify species of the strains.
Phylogenetic trees were constructed using Bayesian analysis (BA). Before the BA, we performed a likelihood ratio test using Modeltest, version 3.7 (Posada and Crandall 1998) to determine the best model under the hierarchical likelihood ratio tests (hLRTs) and Akaike Information Criterion (AIC). Evolutionary best-fit model was selected as the GTR + I + Γ model from a combined five gene data (nr ITS2 and nr partial LSU rDNA, nr SSU rDNA, cp
BA was performed with MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001). Each analysis was initiated from a random starting tree, and the program was set to run four chains of Markov chain Monte Carlo iterations simultaneously for 2,000,000 generations. Trees and parameters were sampled every 1,000 generations, and the burn-in point was identified graphically by tracking the likelihoods (Tracer V.1.5; http://tree.bio.ed.ac.uk/software/tracer/), and then the first 800 trees were burned to ensure that they had stabilized. A majority rule consensus tree was calculated from the remaining trees to examine the posterior probabilities of each clade. The maximum likelihood (ML) phylogenetic analyses were done using the RAxML 7.0.4 program (Stamatakis 2006) with the general time reversible (GTR) model. We used 1,000 independent tree inferences using the -# option of the program to identify the best tree. Bootstrap values were calculated using 1,000 replicates, using the same substitution model.
>
Secondary structure prediction
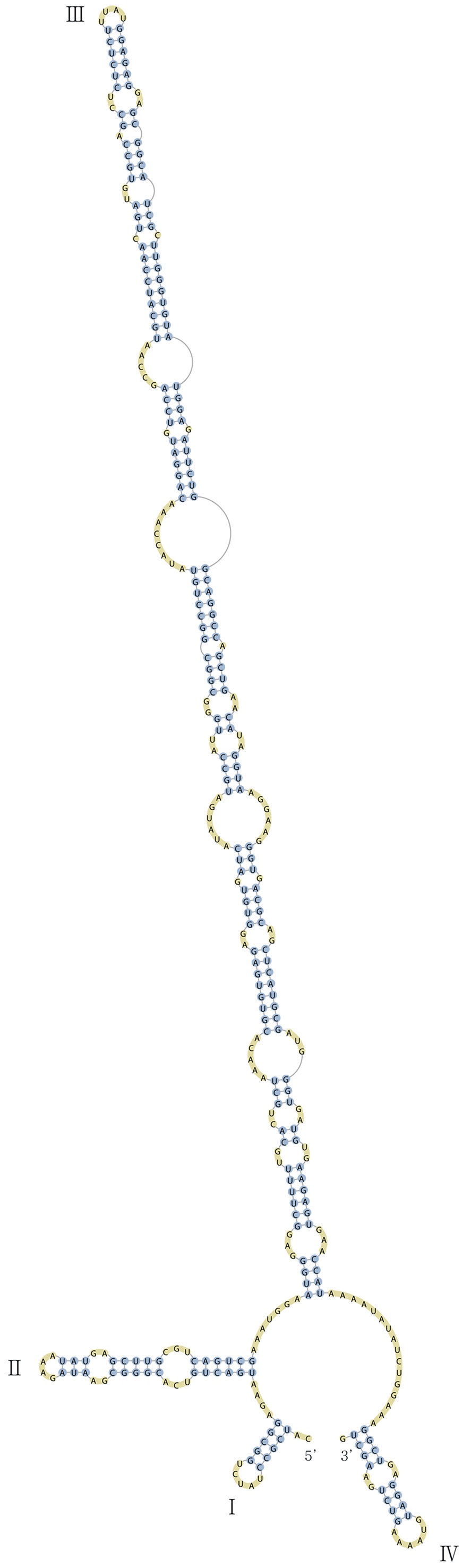
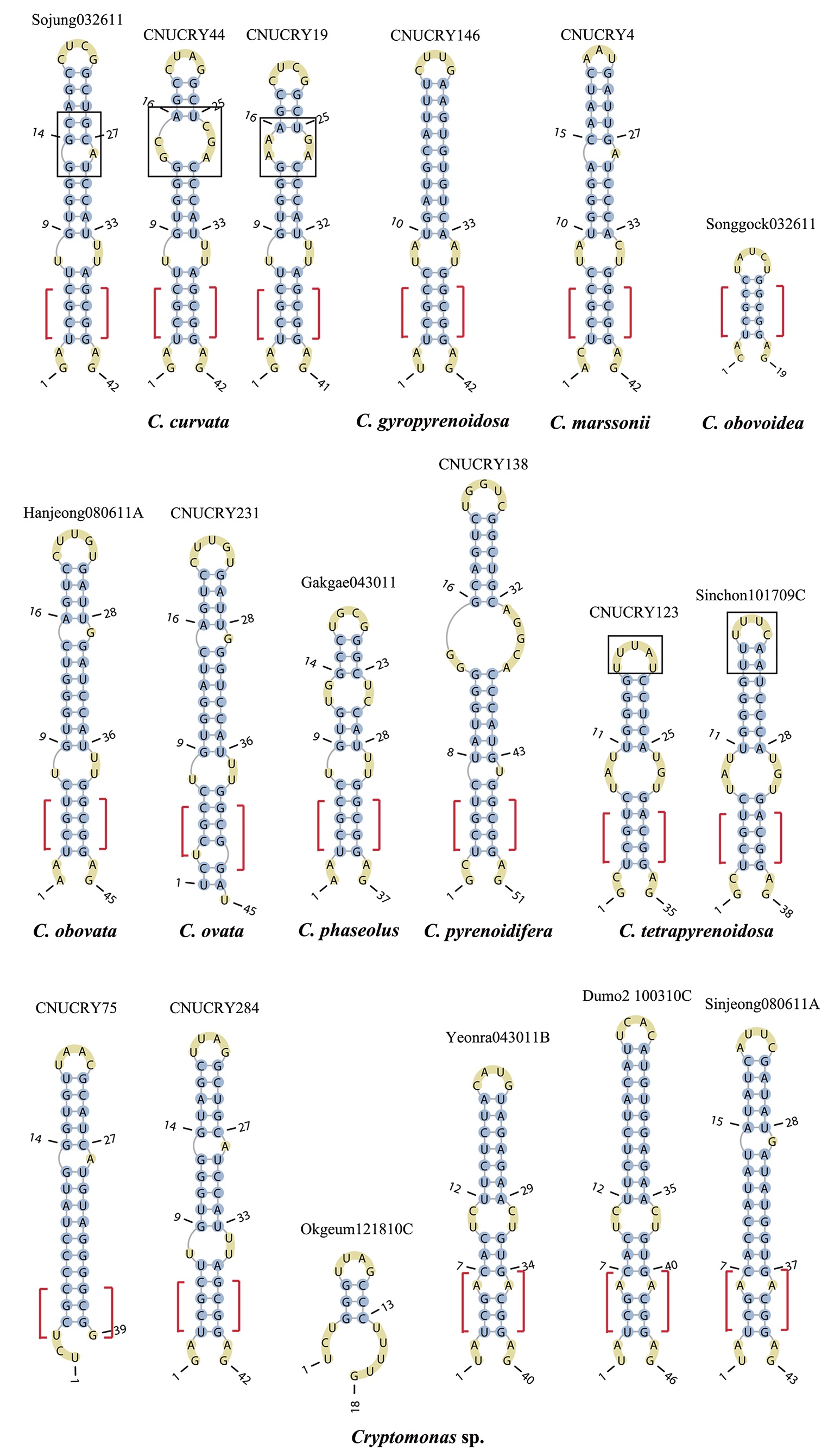
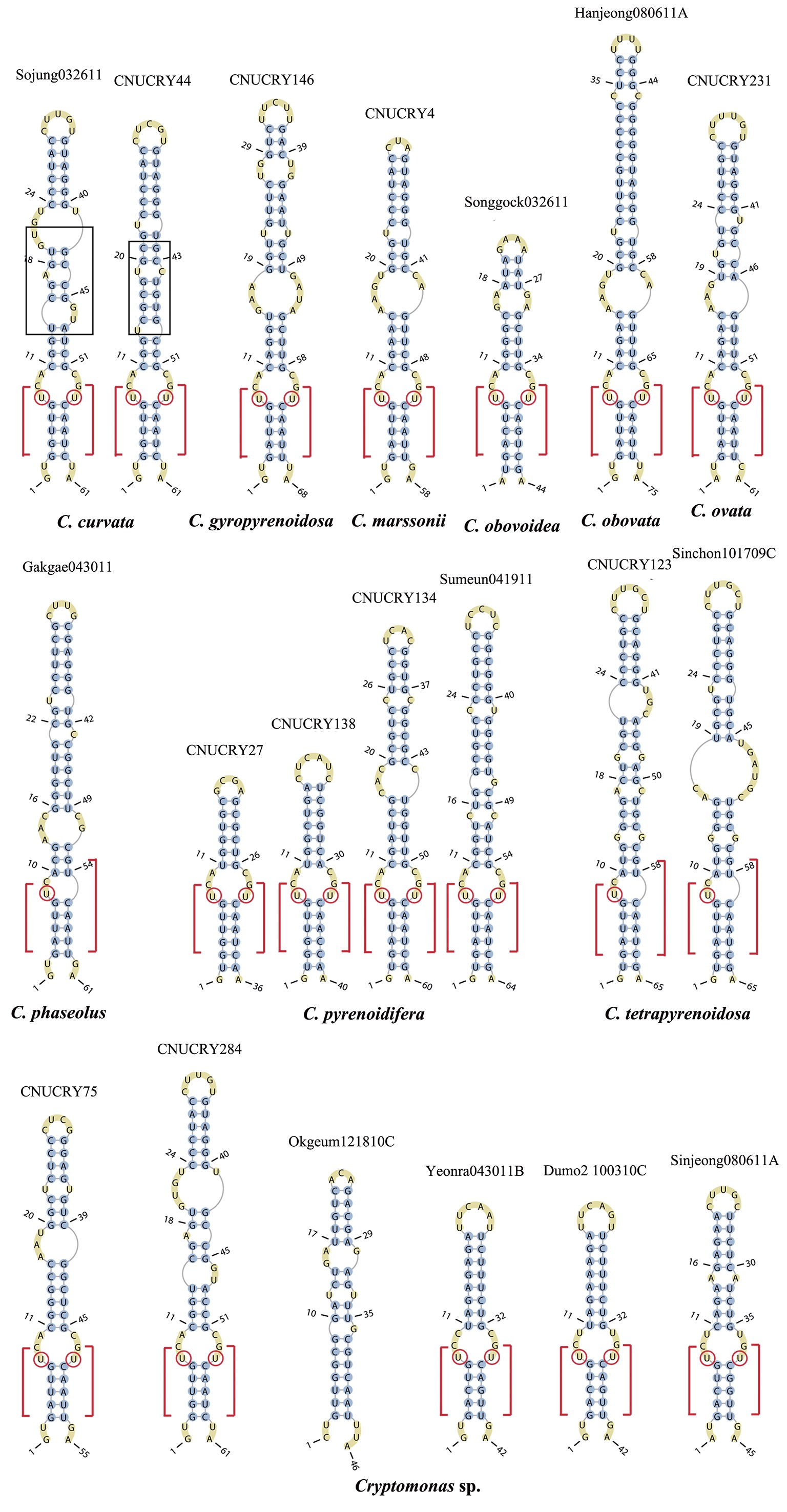
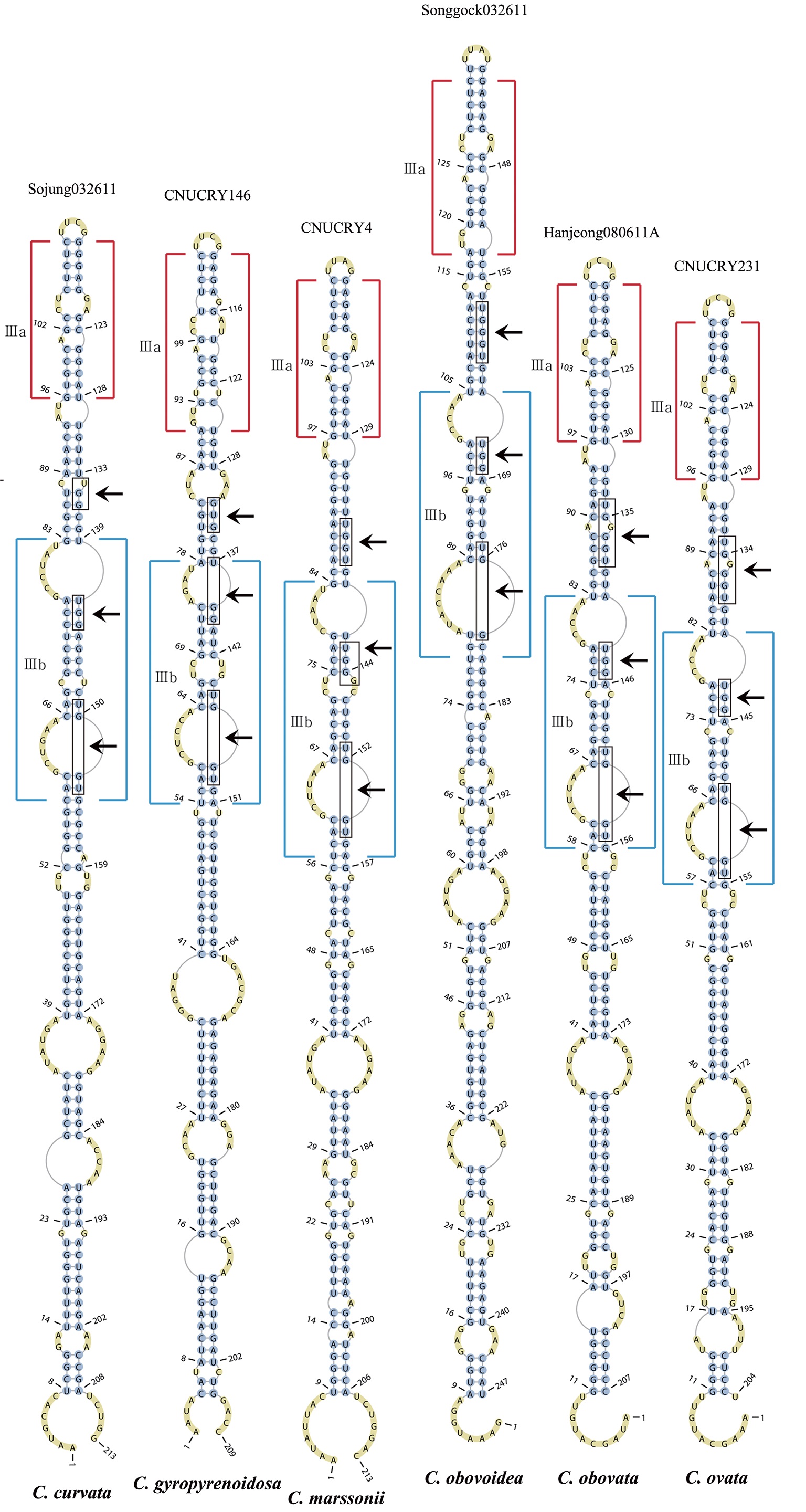
Nuclear ITS2 sequences were folded using the mfold server(http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form) (Zuker 2003) and RNAfold server (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi)(Mathews 2004) with default values. A complete secondary structure graph of the nuclear ITS2 of
Nuclear ITS2, partial LSU rDNA, and SSU rDNA, and chloroplast
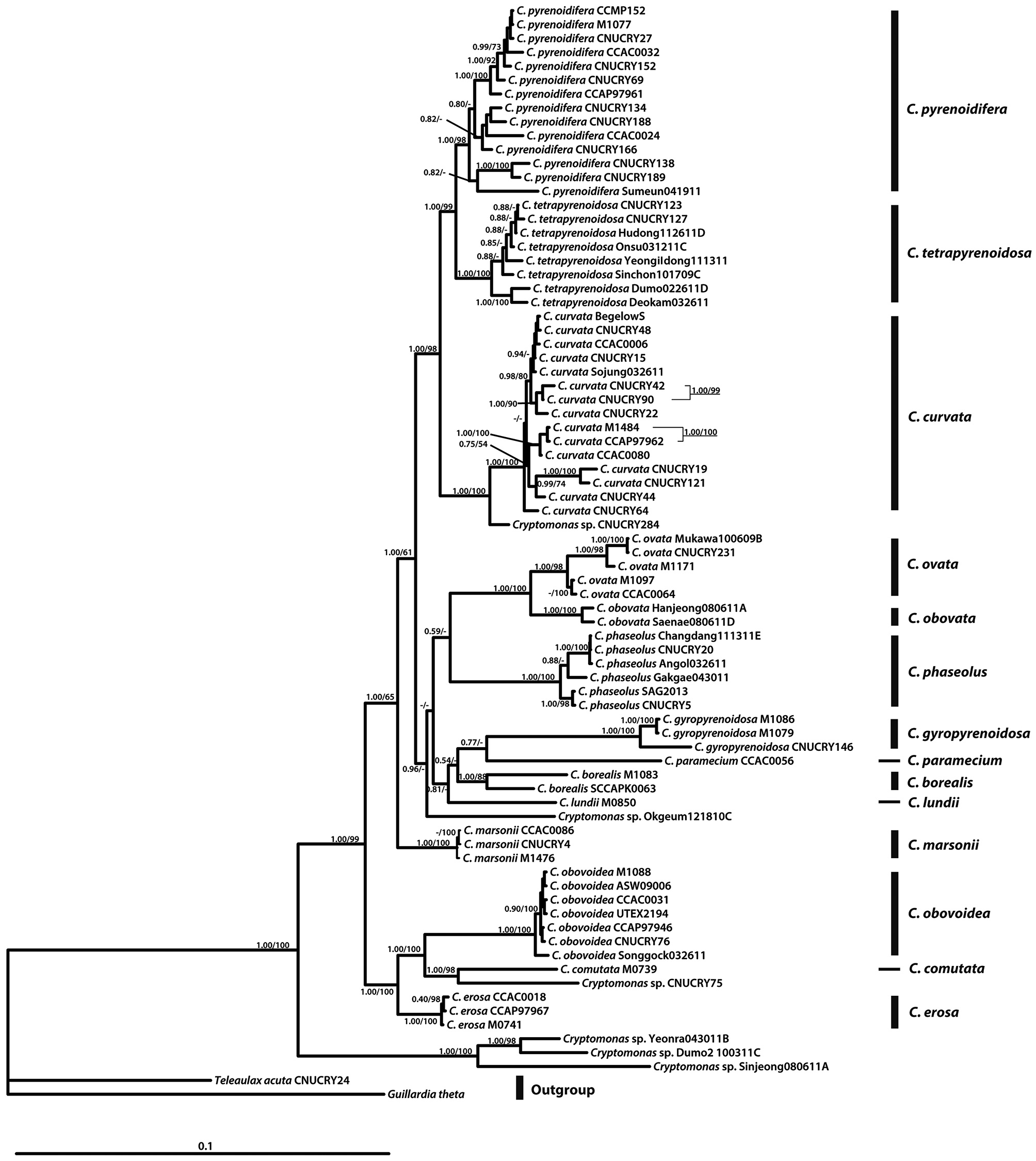
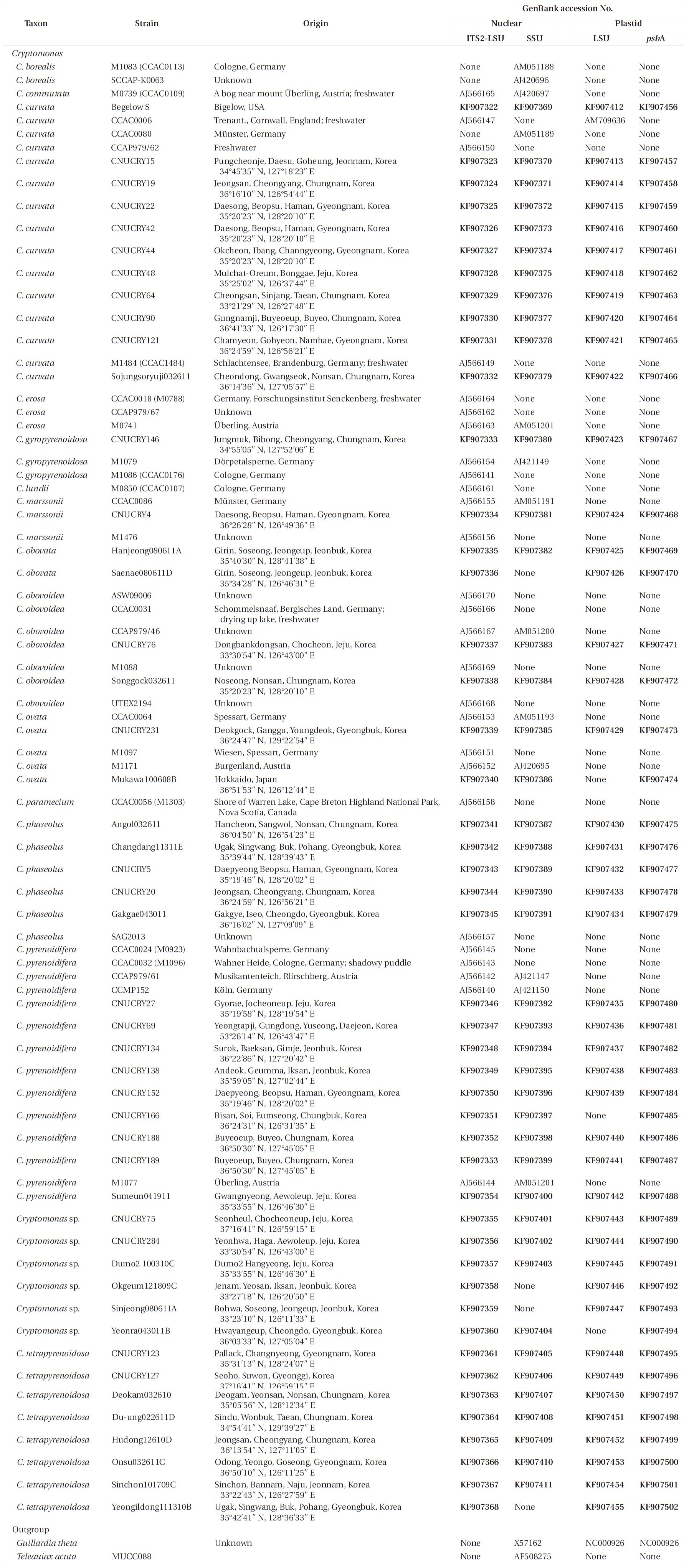
For phylogenetic analyses, 79 strains were used (Table 2). Forty-seven new
>
Secondary structure prediction of nuclear ITS2
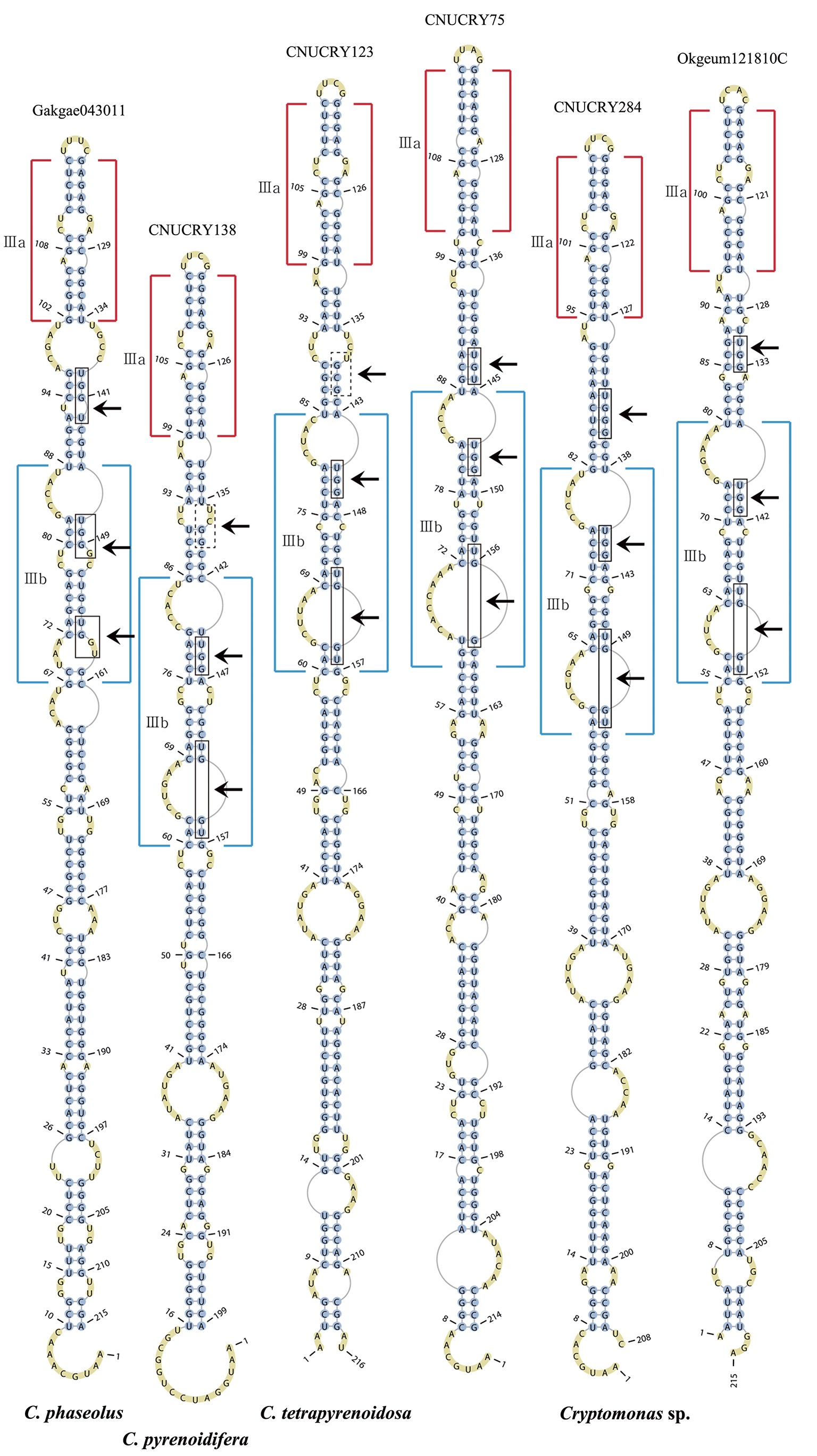
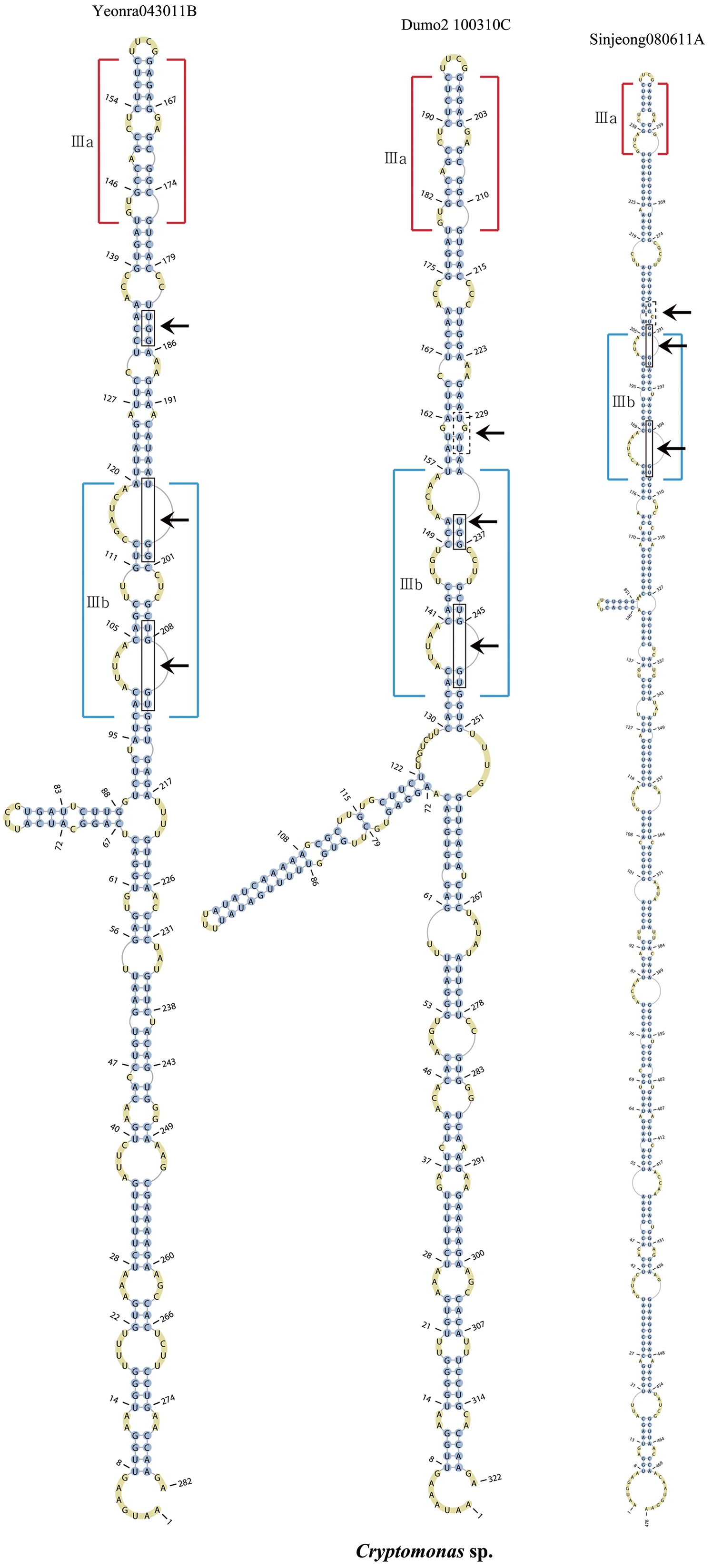
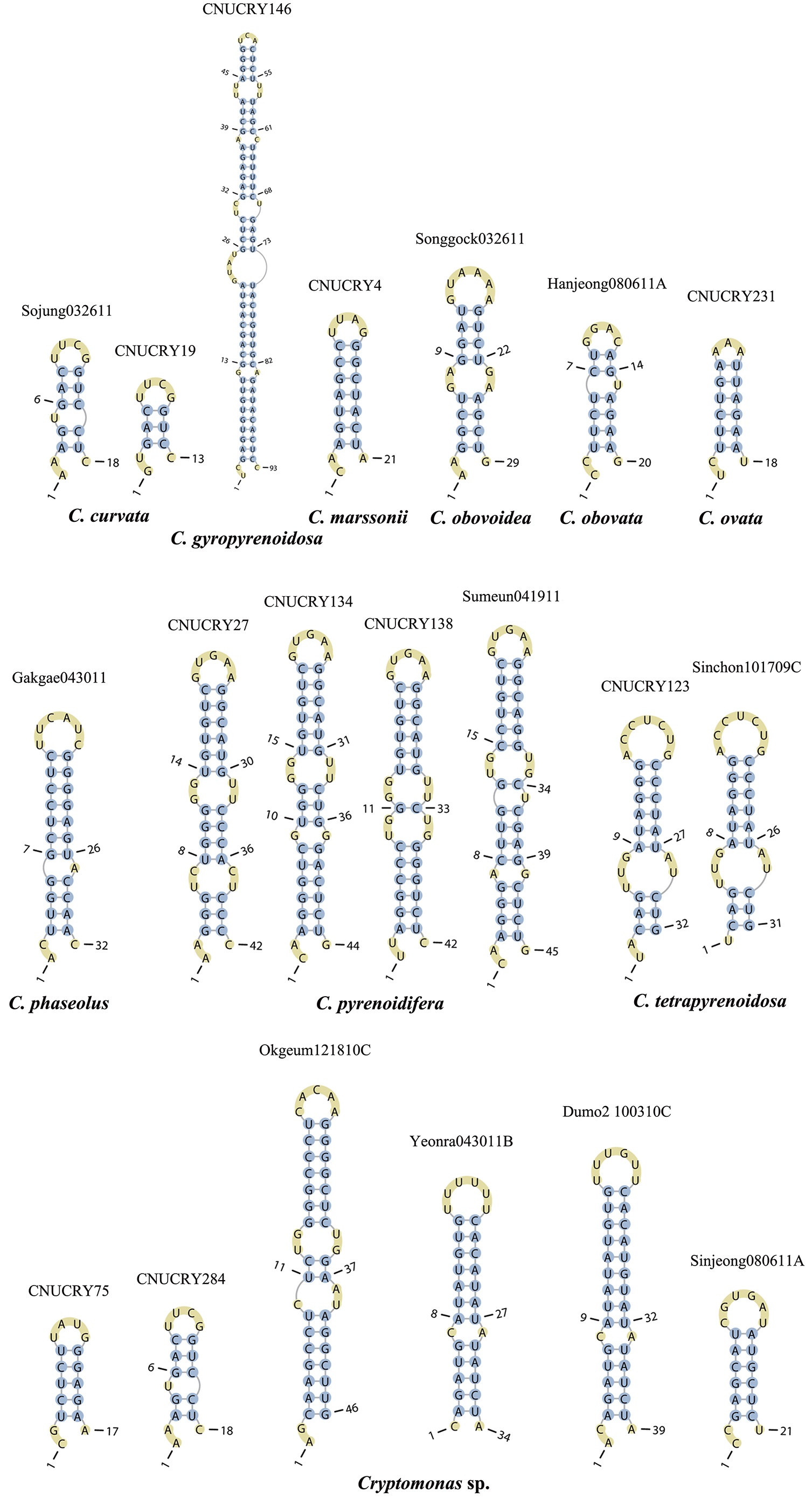
The lengths of most nuclear ITS2 sequences of the examined strains were between 330 (
Across species, the helices I and II were quiet variable in their terminal parts and also varied in length (Figs 4 & 5). The numbers of internal loops differed from zero (
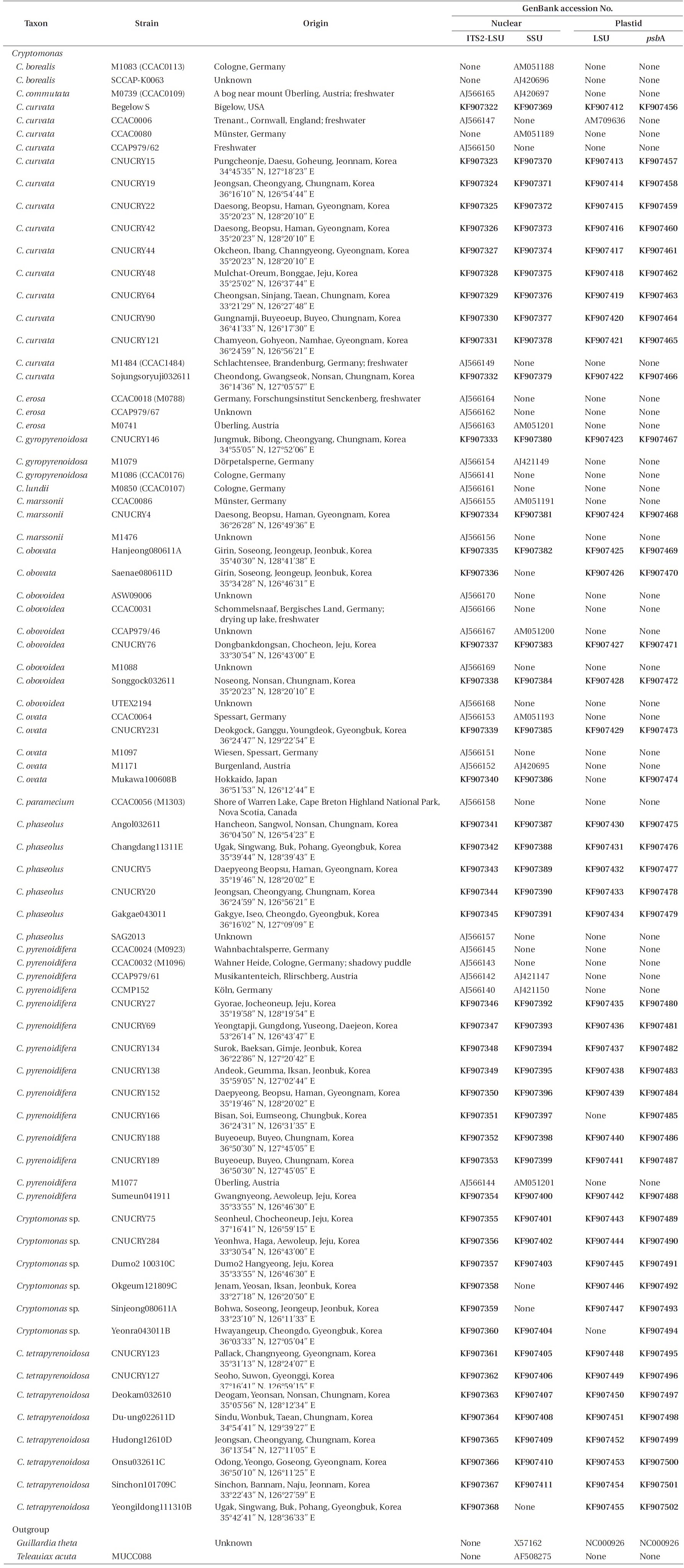
Strains of the genus Cryptomonas used in this study and the GenBank accession numbers for their nr 5.8S-ITS2-LSU, nr SSU, pt psbA and pt LSU rDNA sequences
The shortest helix I was found in
>
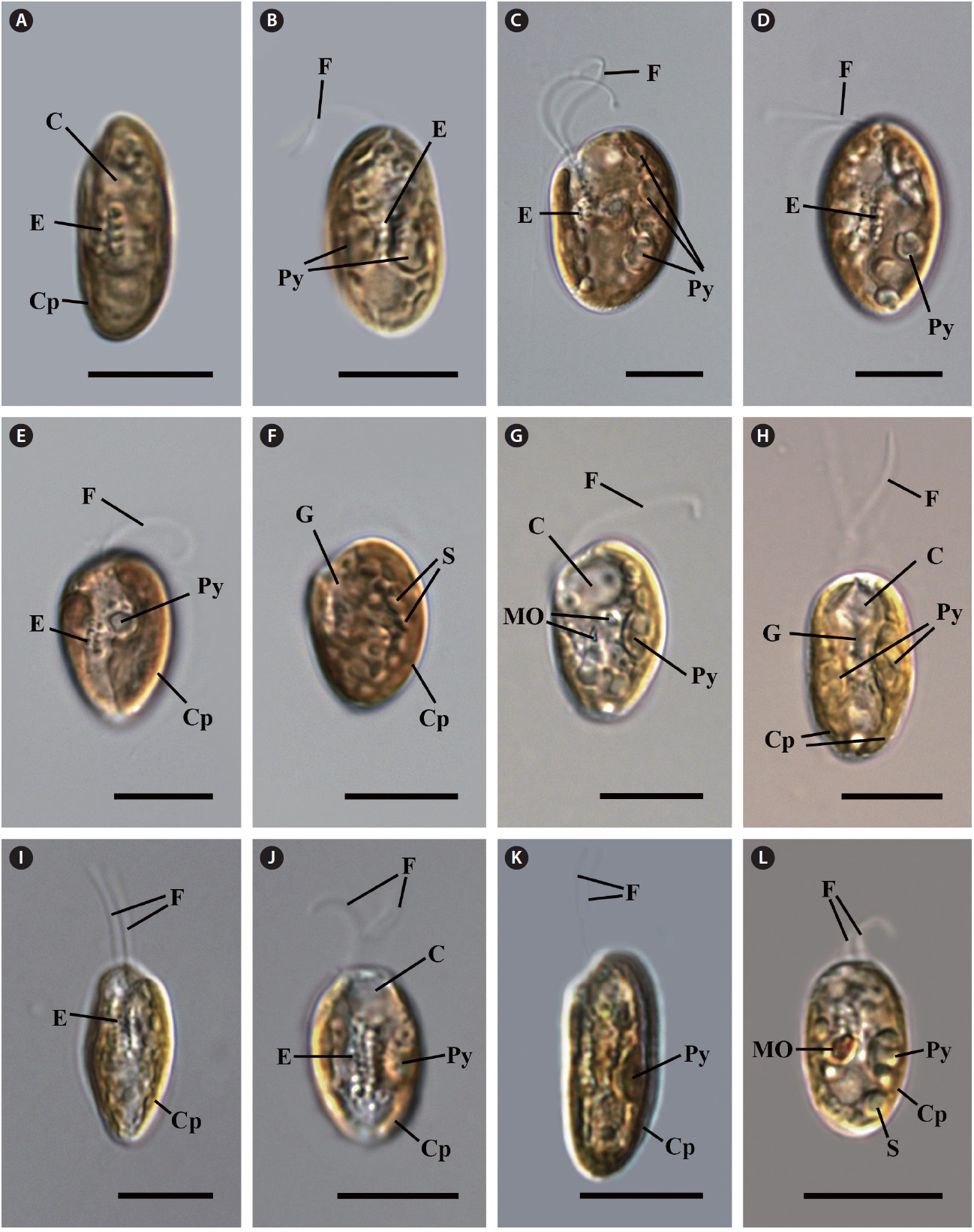
Morphology of Cryptomonas species from Korea
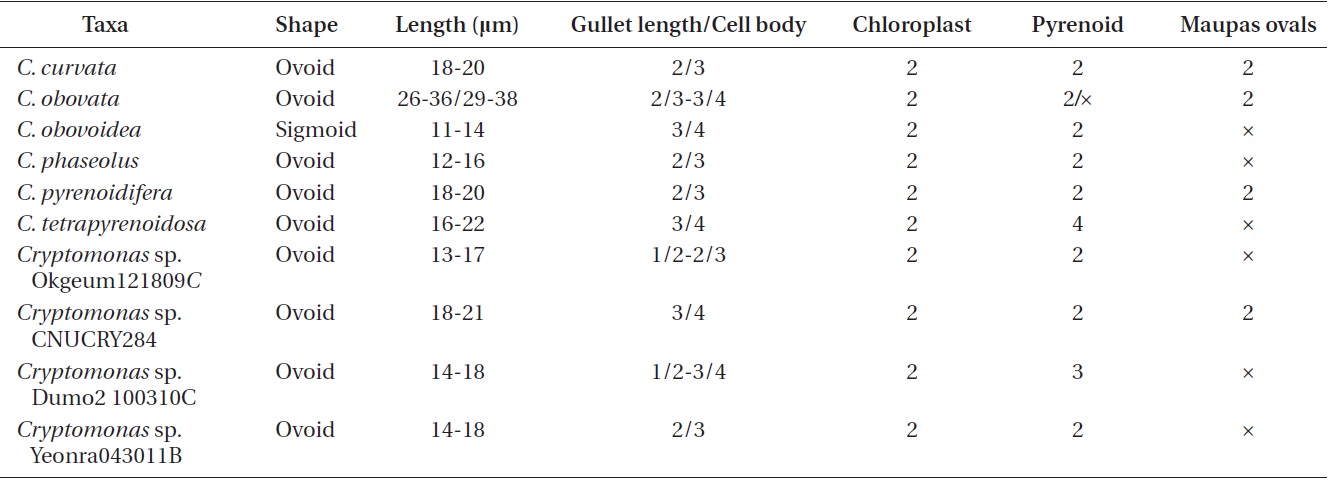
Cell shape and length, length of the fullow-gullet system, color and number of chloroplasts, and the presence / absence and number of pyrenoids were examined. The results are summarized in Table 3 and illustrated in Figs 10-12.
[Table 3.] Morphological points of comparisons among Korean Cryptomonas species
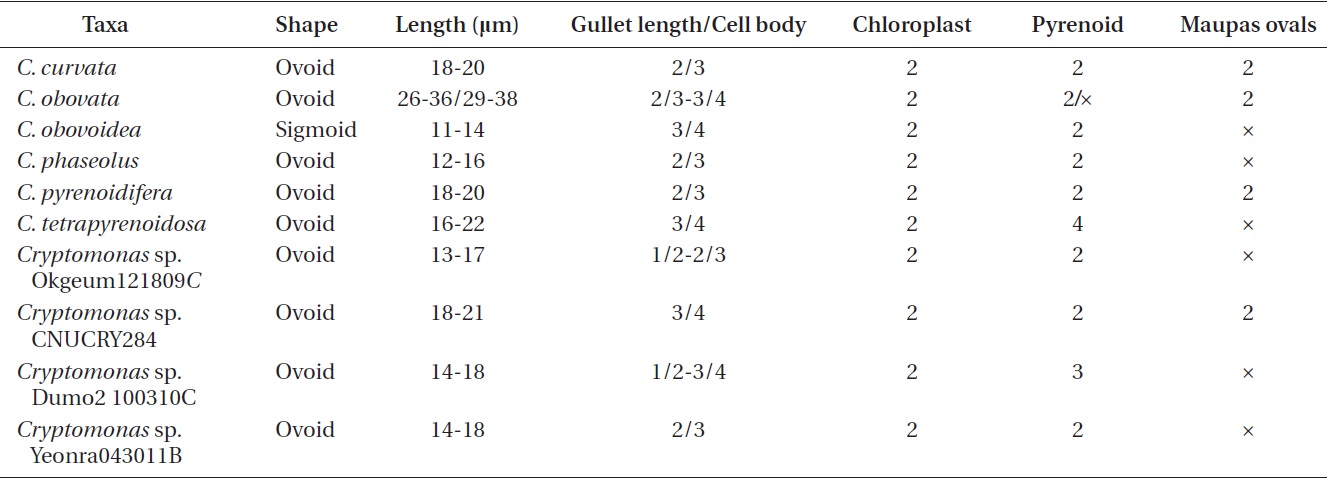
Morphological points of comparisons among Korean Cryptomonas species
>
Cryptomonas curvata (Ehrenberg) Hoef-Emden et Melkonian 2003
Specimens examined. Sojungsoryuji, Namsan, Korea; Seohojeosuji, Suwon, Korea; Daepyeong swamp, Haman, Korea; Jjokji pond, Changyeong, Korea; 1100goji, Seogwipo, Korea; Chungsan, Taean, Korea; Gungnamji, Buyeo, Korea.
Light microscopy.
>
Cryptomonas gyropyrenoidosa Hoef-Emden et Melkonian 2003
Specimens examined. Jungmukji, Cheongyang, Korea.
Light microscopy.
>
Cryptomonas marssonii (Skuja) Hoef-Emden et Melkonian 2003
Specimens examined. Daesong, Haman, Gyeongnam, Korea.
Light microscopy.
>
Cryptomonas ovata (Ehrenberg) Hoef-Emden et Melkonian 2003
Specimens examined. Songcheonji, Iksan, Korea; Mukawa, Muroran, Hokkaido, Japan.
Light microscopy.
>
Cryptomonas obovata Czosnowski 1948
Specimens examined. Saenaebangjuk, Jeongeup, Korea; Hanjeongje, Jeongeup, Korea.
Light microscopy.
>
Cryptomonas obovoidea (Pascher) Hoef-Emden 2007
Specimens examined. Songgock, Nonsan, Korea; Mokmalji, Taean, Korea; Dongbackdongsan, Jeju, Korea.
Light microscopy.
>
Cryptomonas phaseolus (Skuja) Hoef-Emden 2007
Specimens examined. Angolji, Nonsan, Korea; Gakgaeji, Cheongdo, Korea; Jangdangji, Pohang, Korea; Daepyeong swamp, Haman, Korea; Jeongsan, Cheongyang, Korea.
Light microscopy.
>
Cryptomonas pyrenoidifera (Geitler) Hoef-Emden et Melkonian 2003
Specimens examined. Gyorae, Jeju, Korea; Yeongtapji, Daejeon, Korea; Wonseongje, Surok, Baeksan, Korea; Andeokjeosuji, Iksan, Korea; Soiji, Bisan, Eumseong, Korea; Gungnamji, Buyeo, Korea; Sumeunmulbaengdeui, Aewol, Jeju, Korea.
Light microscopy.
>
Cryptomonas tetrapyrenoidosa (Skuja) Hoef-Emden et Melkonian 2003
Specimens examined. Sinchonje, Naju, Korea; Duungseupji, Taean, Korea; Onsusoryuji, Goseong, Korea; Deokamji, Nonsan, Korea; Yeongildongji, Pohang, Korea; Hudongje, Cheongyang, Korea; Seohojeosuji, Suwon, Korea.
Light microscopy.
an ovoid shape in ventral view (Fig. 12C & D). The cells were 16-22 μm in length. The length of gullet with rows of ejectisomes was 2/3 that of cells. Cells had two brownchloroplasts, and each chloroplast had two or three pyrenoids facing each other in the left and right chloroplast lobes. Contractile vacuole was located in apical and two flagella inserted in a vestibulum of the cell invagination. Cells had two maupas ovals.
>
Cryptomonas sp. Okgeumji121810C
Specimens examined. Okgeumji, Iksan, Korea.
Light microscopy.
Specimens examined. Yeonhwaji, Jeju, Korea.
Light microscopy.
>
Cryptomonas sp. Dumo2 100310C
Specimens examined. Dumojeosuji, Jeju, Korea.
Light microscopy.
>
Cryptomonas sp. Yeonra043011B
Specimens examined. Yeonra, Hwayangeup, Cheongdo, Gyeongbuk, Korea.
Light microscopy.
Although cryptomonad genera were delimited by ultrastructral features, such as periplast and flagellar apparatus, all
>
Molecular phylogenetic analyses
In the molecular phylogenetic tree based on nuclear 18S rDNA, the genus
>
Nuclear ITS2 secondary structures
The nuclear ITS2 in
The helix ?, ??, and ???b were less conserved regions than helix ???a (the distal part of helix ???). The proximal part of helix ? and ?? had similar sequences among species, but were variable in their terminal parts. Specific structures of helix ???b appeared in all strains, despite the differences among sequences. Sequences of helix ?, ??, and ???b in ITS2 secondary structure were very useful for the identification of