The unarmored dinoflagellates, Amphidinium spp. Claperede and Lachmann 1859, are characterized by a small epicone relative to total cell size. They are ubiquitous and many of them have been found in subtropical or tropical waters (Murray and Patterson 2002). In addition, they live in diverse habitats, such as pelagic, salt marsh, and benthic environments (Dodge 1982, Larsen 1985, Larsen and Patterson 1990, Jørgensen et al. 2004, Dolapsakis and Economou-Amilli 2009). Furthermore, most species are free-living forms, although some are endosymbiotic (Taylor 1971, Lopes and Silveira 1994, Murray et al. 2004). In trophic modes, some species are phototrophic, while other species are heterotrophic (Baillie 1971, Larsen 1988, Calado et al. 1998, Jørgensen et al. 2004). Some Amphidinium species produce toxins, such as ciguatera fish poisoning or hemolysins, and have caused mass fish mortality (Yasumoto 1990, Hallegraeff 1993, Nayak et al. 1997, Baig et al. 2006, Selina and Levchenko 2011). Thus, identifying Amphidinium species is of critical concern to scientists and people involved in aquaculture industries. However, due to the ambiguity of Amphidinium taxonomy, the classification of species has been re-examined (Jørgensen et al. 2004, Murray et al. 2004, 2012).
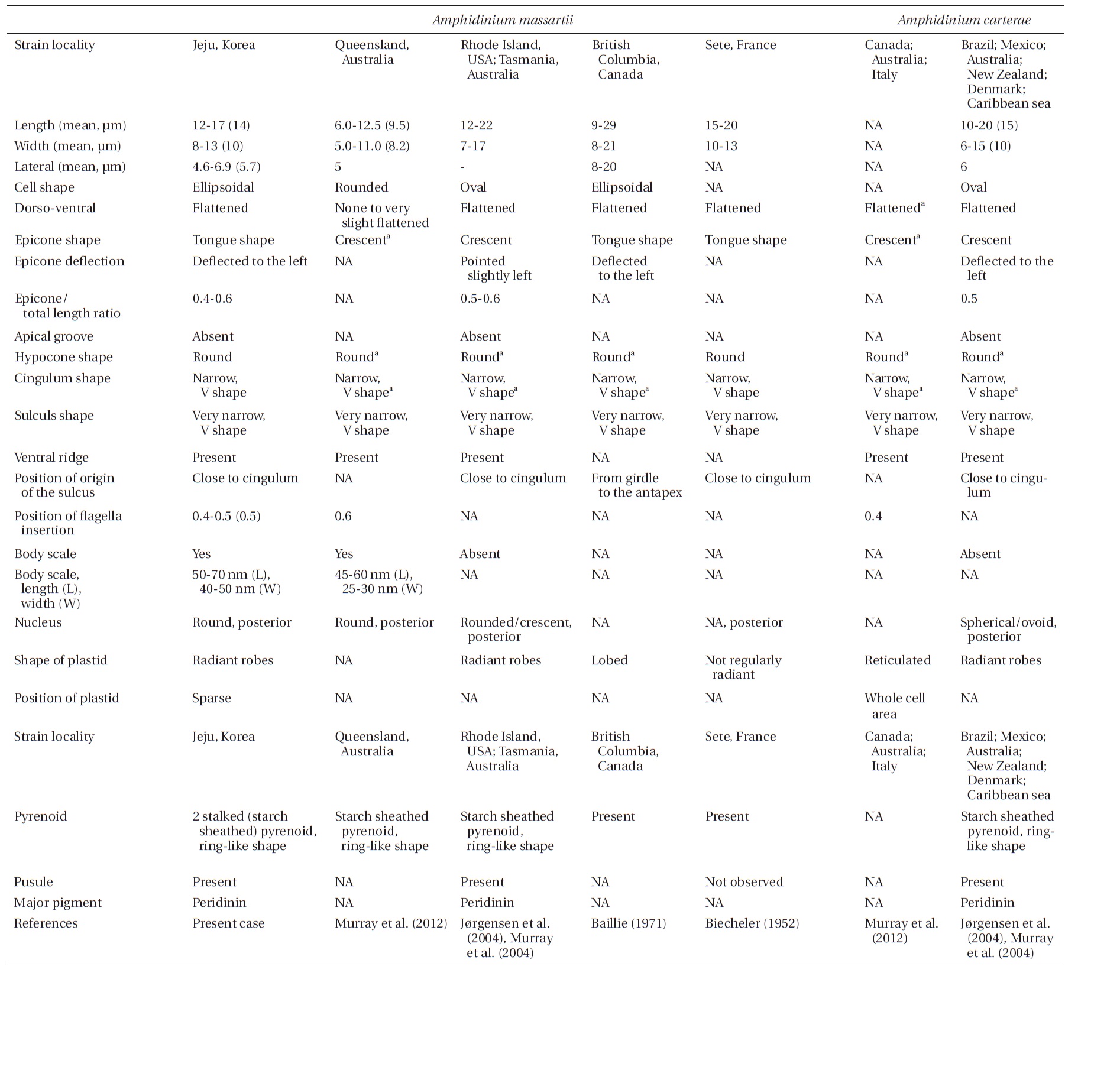
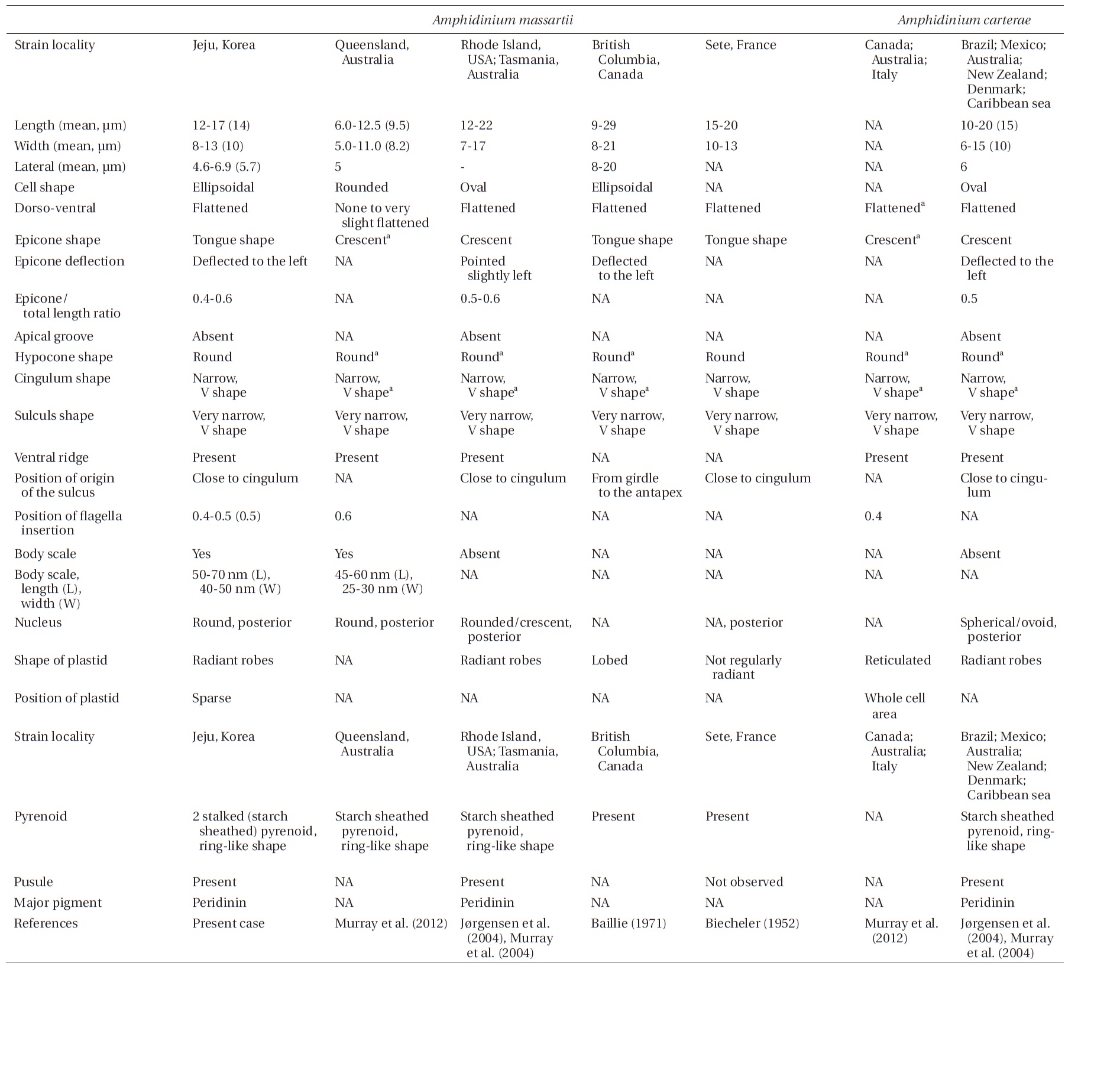
Amphidinium massartii Biecheler (1952) and Amphidinium carterae Hulburt (1957) are small and have sometimes been considered to be con-specific (Murray et al. 2004, 2012). However, Murray et al. (2004) suggested that A. massartii is clearly different from A. carterae because A. massartii plastids are centrally located and are present at a relatively low density, while A. carterae plastids are peripherally located and reticulated. In addition, they also suggested that flagellar insertion of A. massartii (~0.6 of the cell body from the apex) is situated in a lower position than in A. carterae (~0.4 of the cell body from the apex). Furthermore, they showed that in phylogenetic trees based on internal transcribed spacer (ITS) and large subunit (LSU) rDNA sequences and of cytochrome b sequences, the A. massartii clade is clearly divergent from the A. carterae clade.
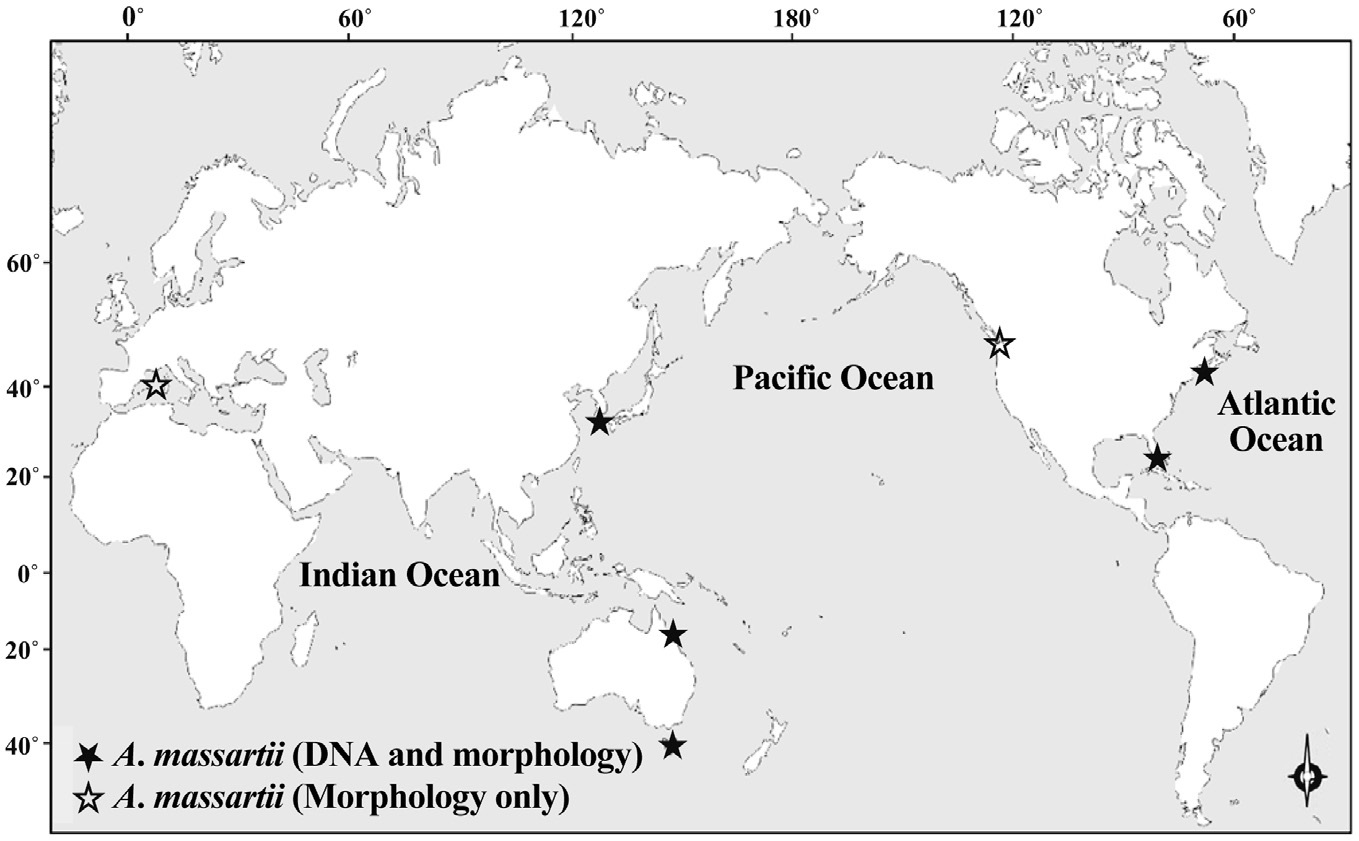
While there have been many studies of the taxonomy and ecology of A. carterae, which has a worldwide distribution, there have been few studies of the taxonomy and ecology of A. massartii (Jørgensen et al. 2004, Murray et al. 2004, 2012). A. massartii has so far been reported in the waters of Australia, USA, Canada, and southern France (Biecheler 1952, Baillie 1971, Jørgensen et al. 2004, Murray et al. 2004, 2012). Therefore, it is worthwhile exploring the presence of this species in other waters.
Recently, we isolated Amphidinium cells from temperate waters near seaweed beds off of Jeju Island, Korea and established 3 clonal cultures. We then conducted morphological and genetic analyses using these cultures. The morphology and rDNA sequences of these strains were very similar to those of A. massartii (Murray et al. 2012). Therefore, in the present study, we report the morphological and genetic characteristics of Korean strains of A. massartii, and present phylogenetic trees based on small subunit (SSU), ITS1, 5.8S, ITS2, and LSU rDNA sequences. The present study provides a basis for understanding the distribution of A. massartii in the world and the intraspecific and interspecific variations in the morphology and DNA sequences of Amphidinium spp.
For the isolation and culture of A. massartii, samples of articulated coralline algae were collected by divers at a depth of ~2 m off Namwon, Jeju Island, Korea (33°3′ N, 126°7′ E). Algae were collected in October 2011 when water temperature and salinity were 22.7℃ and 33.7, respectively. The samples were placed in plastic bags and transported to the laboratory. Samples were then vigorously shaken to detach the dinoflagellates from the algae, screened through a 300-μm Nitex mesh, and placed in six-well tissue culture plates. Each of three clonal cultures of A. massartii was established by two serial single-cell isolations. Bottles containing f/2 medium and A. massartii were filled to capacity with freshly filtered seawater, capped, and placed on a shelf at 20℃ under illumination by cool white fluorescent lights at 20 μmol photons m-2 s-1 under a 14 : 10 h light-dark cycle. As the concentration of A. massartii increased, A. massartii was subsequently transferred to 50-, 125-, and 500-mL polycarbonate (PC) bottles containing fresh f/2 seawater medium. Once dense cultures of A. massartii were obtained, they were transferred approximately every 5 weeks to new 500-mL PC bottles containing fresh f/2 seawater medium. When the concentration of A. massartii became high, we analyzed the DNA sequences of the cultured cells. After genetic identification, and when culture volumes were sufficiently large, the morphology of the dinoflagellates was analyzed. Only cultures in the exponential growth phase (i.e., growth rate = ~0.1 d-1) were used.
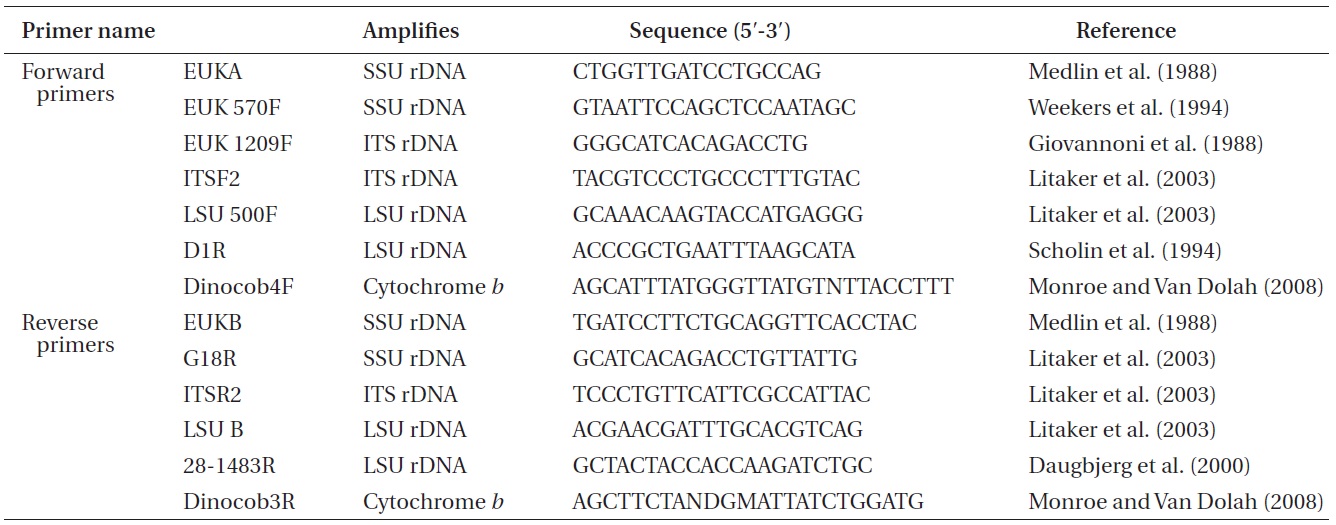
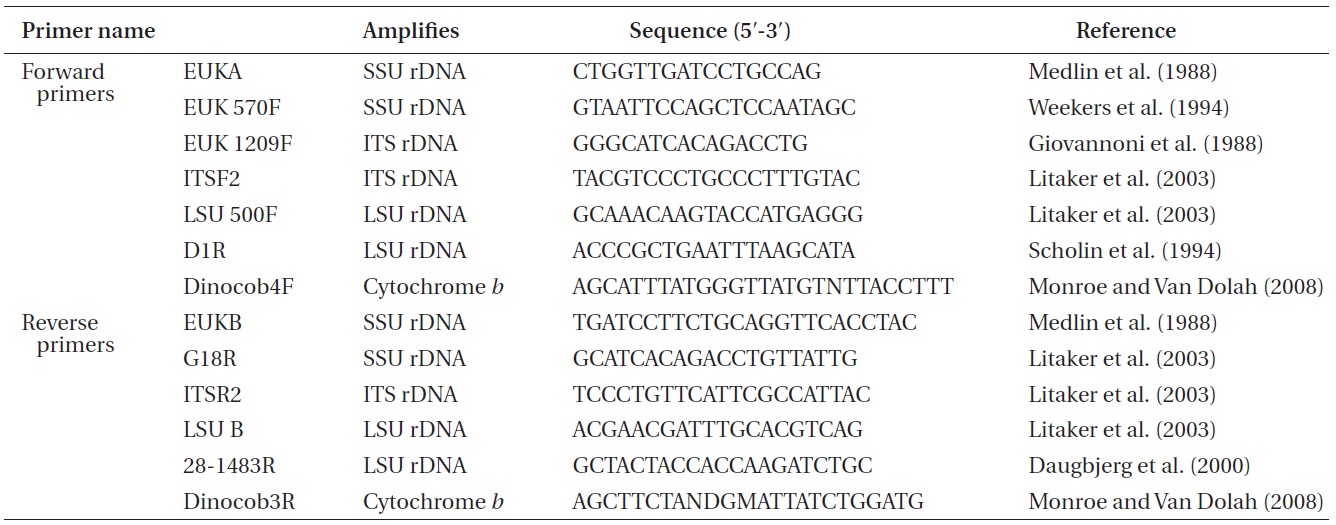
Approximately 10-mL of a dense culture of A. massartii was concentrated by centrifugation at 2,190 ×g for 15 min at room temperature, and the pellet was used for genomic DNA extraction. The genomic DNA extraction; amplification of the SSU, ITS1, 5.8S, ITS2, and LSU rDNA; and PCR reactions, sequencing, and alignment were as in Kang et al. (2011). In addition, cytochrome b, one of the mitochondrial genes, was also analyzed using the methods for rDNA analysis described by Kang et al. (2011), except that different primers were used (Table 1).
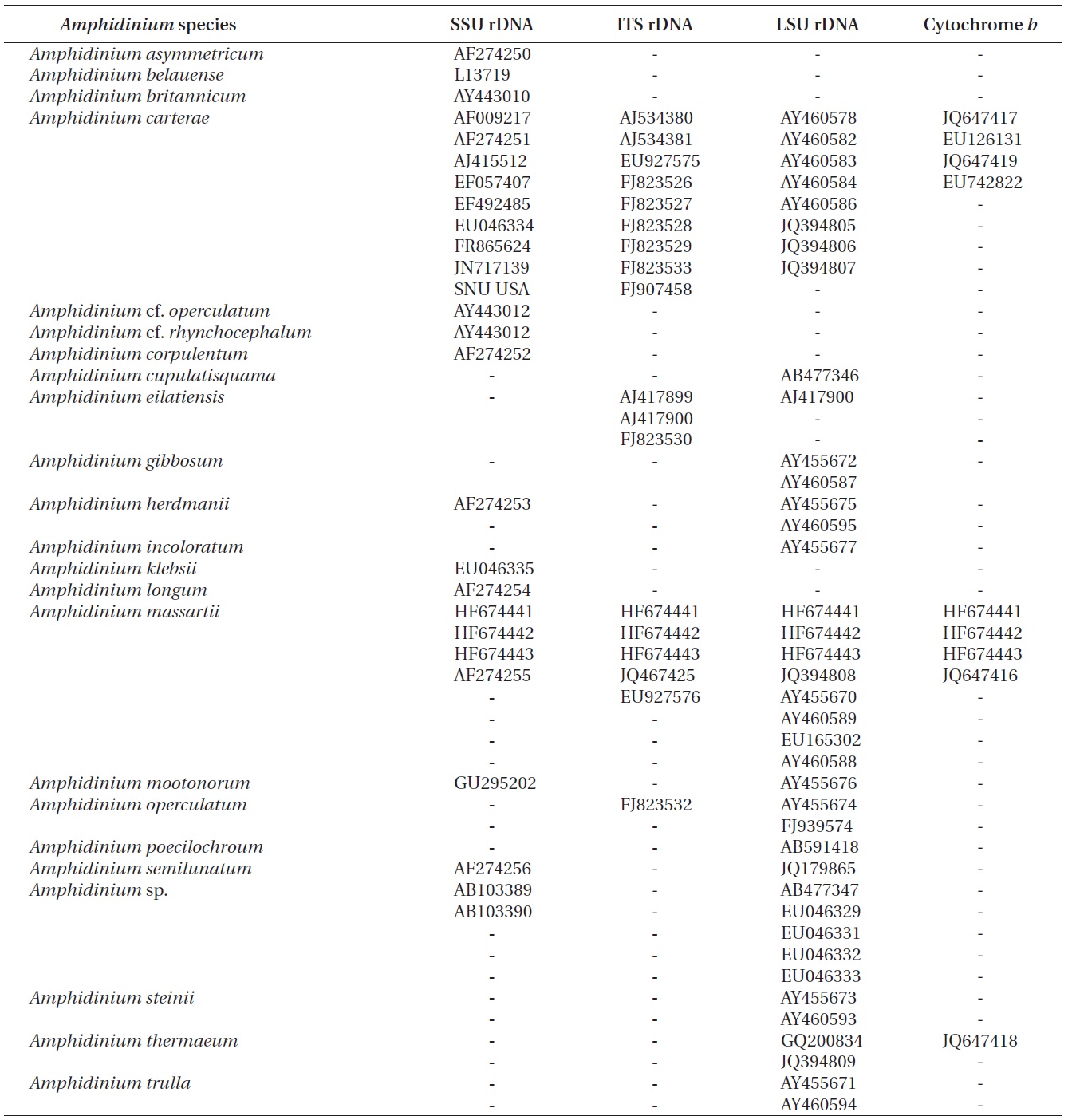
Nuclear SSU, ITS, and LSU rDNA sequences of Amphidinium spp. (Table 2) were aligned using MEGA v. 4 (Tamura et al. 2007) and CLUSTAL X2 (Larkin et al. 2007), and double-checked using the Genetic Data Environment (GDE 2.2) program (Smith et al. 1994) with diverse assemblages of other dinoflagellates that are available in GenBank. The analysis of cytochrome b from Amphidinium species and other dinoflagellates was also performed using the same procedures. Maximum likelihood (ML) analysis of each region was conducted using the RAxML 7.0.3 program (Stamatakis 2006) and the default algorithm, a general time reversible (GTR) + ? model. We allowed for 200 independent free inferences using the -# option included in the program in order to identify the best tree. Maximum likihood bootstrap values were calculated using 1,000 replicates under the same substitution model. Bayesian analyses were performed using MrBayes v. 3.1 (Huelsenbeck and Ronquist 2001, Ronquist and Huelsenbeck 2003) in order to determine the best available model for the data from each region. The selected model was GTR + I + G for the SSU, ITS, and LSU rDNA and cytocrhome b region. Four independent Markov chain Monte Carlo simulations (MCMC) were run simultaneously for 2,000,000 generations, and trees were sampled every 1,000 generations. The first 800 trees were deleted to ensure that the likelihood had reached convergence.
The morphology of living cells that were growing photosynthetically was examined using an inverted microscope. The length and width of live cells were measured with the aid of a digital camera (Zeiss AxioCam MRc5; Carl Zeiss Ltd., Gottingen, Germany). For scanning electron microscope, 10-mL aliquots of cultures at ~8,000 cells mL-1 were fixed for 20 min in osmium tetroxide at a final concentration of 2-2.5% (v/v) in seawater. Cell collection, dehydration, drying, and observation were as described by Kang et al. (2010). For transmission electron microscopy, the same procedures were conducted as described by Kang et al. (2010).
We analyzed pigments from A. massartii using highperformance liquid chromatography (LC-10A system; Shimadzu Co., Kyoto, Japan) as in Zapata et al. (2000).
A dense culture of 50,000 cells mL-1 of A. massartii growing photosynthetically in f/2 media and under a 14 : 10 h light-dark cycle under cool white fluorescent light at 20 μmol photons m-2 s-1 was filtered onto a 1.2-μm poresized GF/C filter. Three milliliters of 95% methanol were used for extraction and a Waters C8 column (150 × 1.6 mm, 3.5 μm particle size, 0.01 μm pore size; Waters Corp., Milford, MA, USA) was used for separation. Pigments were identified by retention times and absorption spectra that were identical to those of authentic standards. Pigments were quantified against standards purchased from DHI Water & Environment (Hørsholm, Denmark).
To test the toxicity of A. massartii, we conducted a bioassay using larvae of the brine shrimp Artemia salina.
Encysted eggs of A. salina were hatched in 500-mL of natural seawater under artificial light at 20℃ for 48 h. Ten individual A. salina were placed in each chamber of a 6-well culture plate. The chambers in the plate contained 0, 100, 500, 1,000, 5,000, or 10,000 cells mL-1 of A. massartii AMJJ1. Experimental wells containing both nauplii and dinoflagellates, nauplius-only control wells, dinoflagellate-only control wells, and A. massartii-only control wells were established. All experimental conditions were reproduced in triplicate. The plate chambers were placed on a shelf and incubated at 20℃ under a 14 : 10 h lightdark cycle of cool white fluorescent lights at 20 μmol photons m-2 s-1. At the beginning of incubation and 6, 12, 24, and 48 h later, the plate chambers were placed under a dissecting microscope, and living and dead nauplii were counted at a magnification of ×7-40.
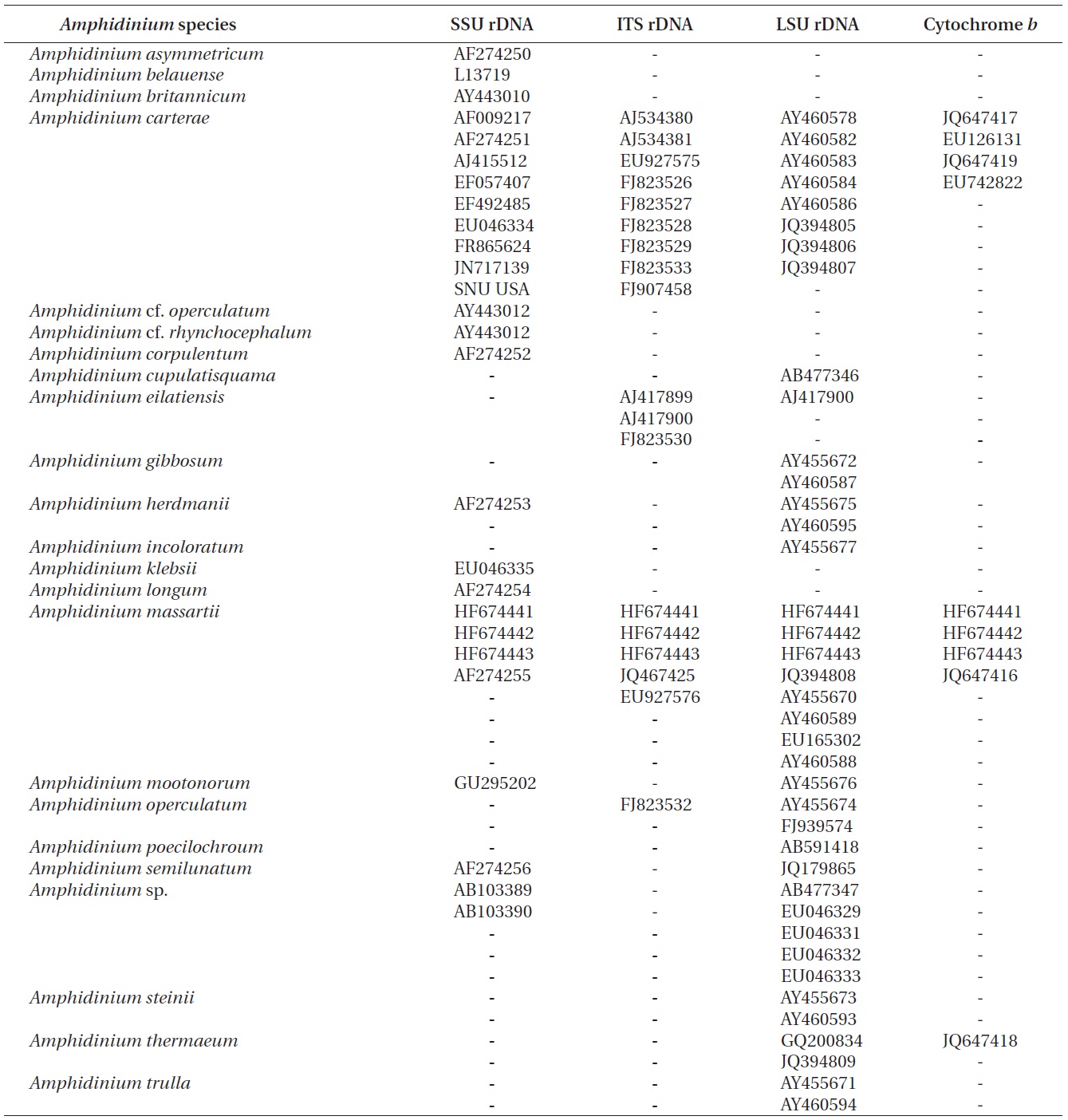
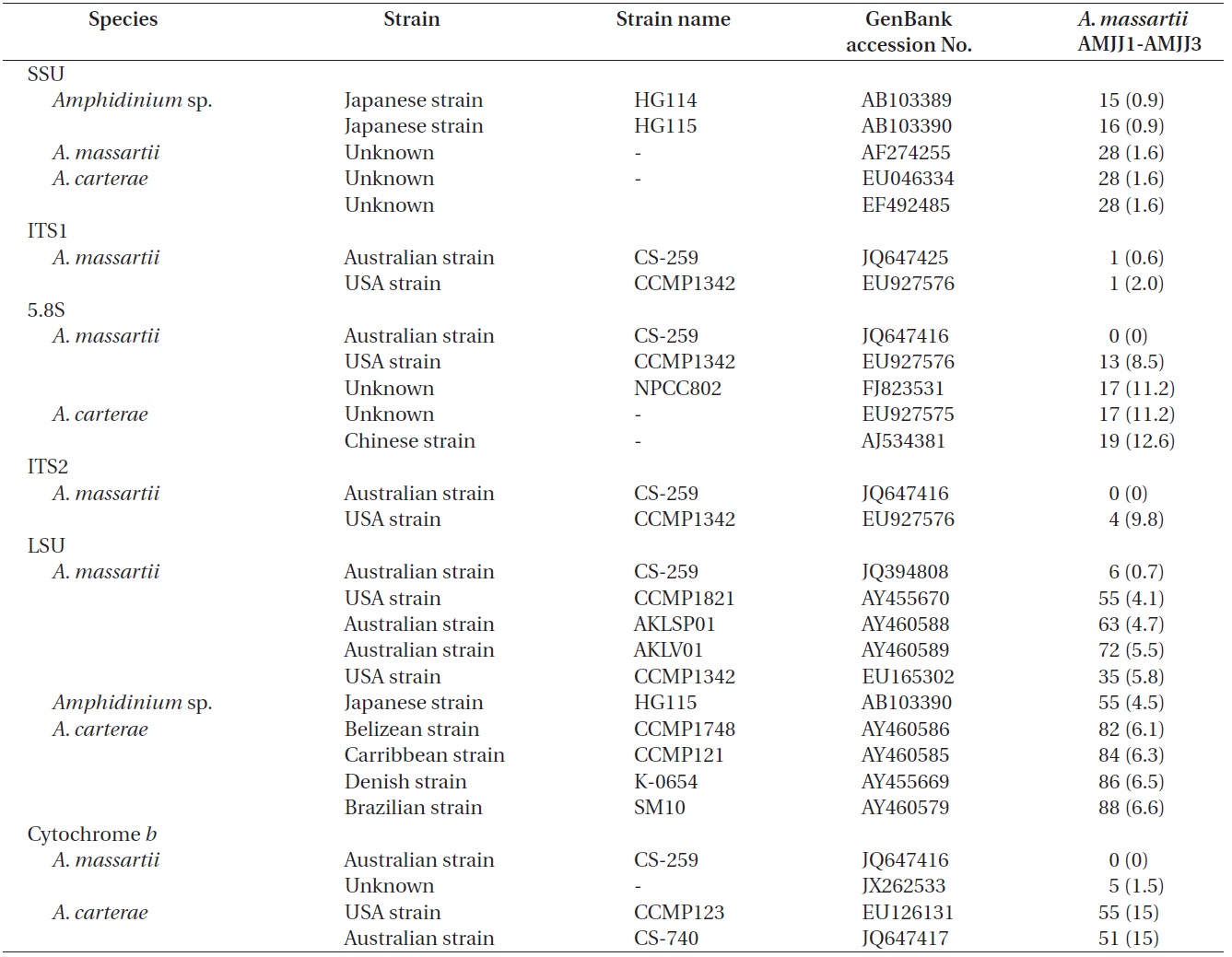
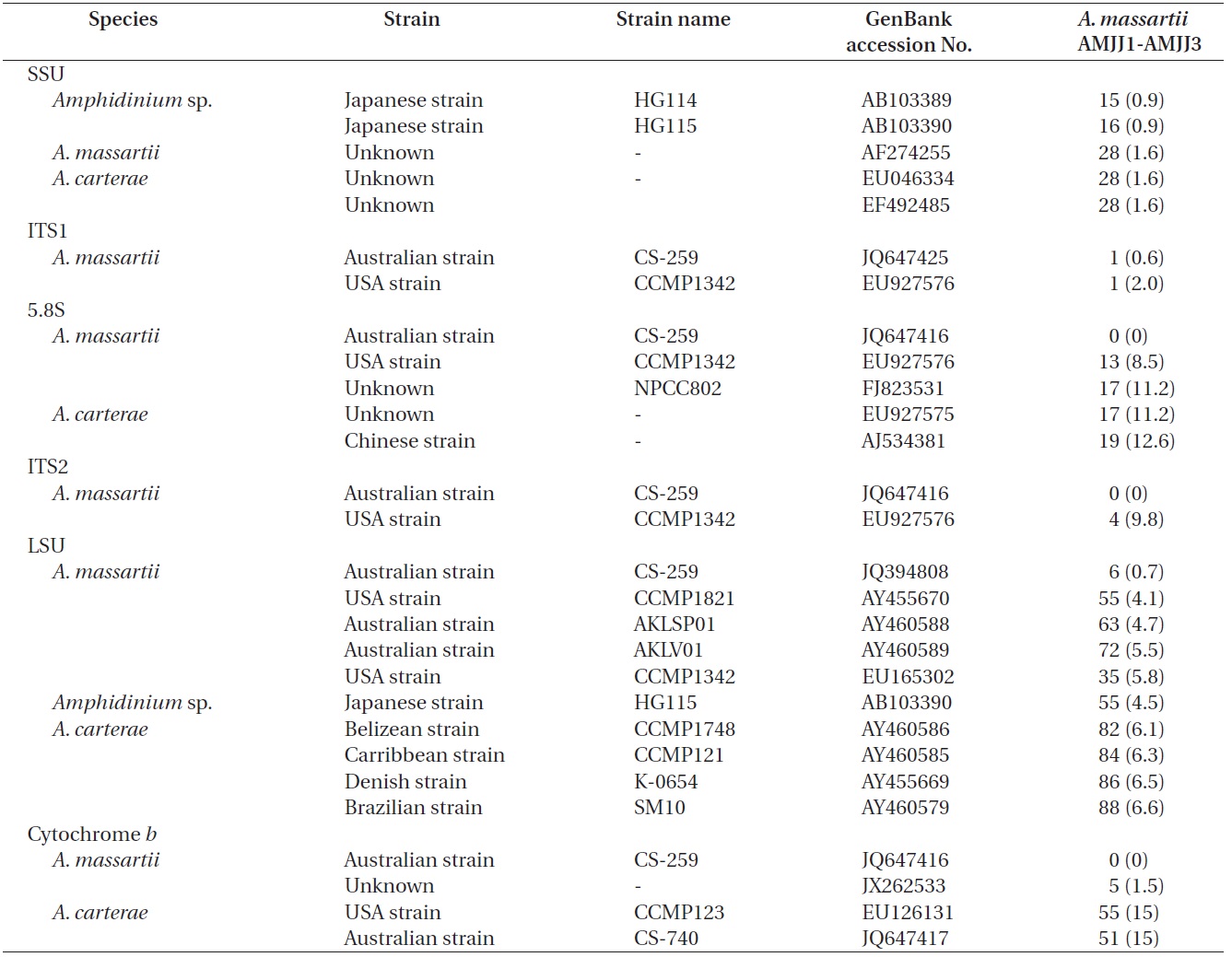
The rDNA sequences of all three Korean strains (Gen- Bank accession number HF674441-674443), 3,575-3,581 bp, were identical. After proper alignment, the LSU rDNA sequences of the Korean strains were 0.7% different from those of an Australian strain of A. massartii CS-259, the closest species, but were 4.1-5.8% different from those of the other Australian and USA strains of A. massartii and those of Amphidinium sp. HG115 that was isolated from subtropical Okinawan waters. The morphology of Amphidinium sp. HG115 has not been formally reported (Table 3). In addition, the ITS1 sequences of the Korean strains were 0.6 and 2.0% different from those of Australian strain CS-259 and USA strains of A. massartii, respectively. The 5.8S rDNA and ITS2 sequences of the Korean strains were identical to those of A. massartii CS-259, but
>8% different from those of a USA strain of A. massartii (CCMP 1342). The SSU rDNA sequences of the Korean strains were 0.9% different from those of Amphidinium sp. HG114 and HG115, which were isolated from subtropical Okinawan waters and 1.6% different from those of an A. massartii strain obtained from an unknown sampling location (Table 3). Furthermore, cytochrome b gene sequences (348-359 bp) of the Korean strains were identical to that of A. massartii CS-259, but >1.5% different from other strains (Table 3).
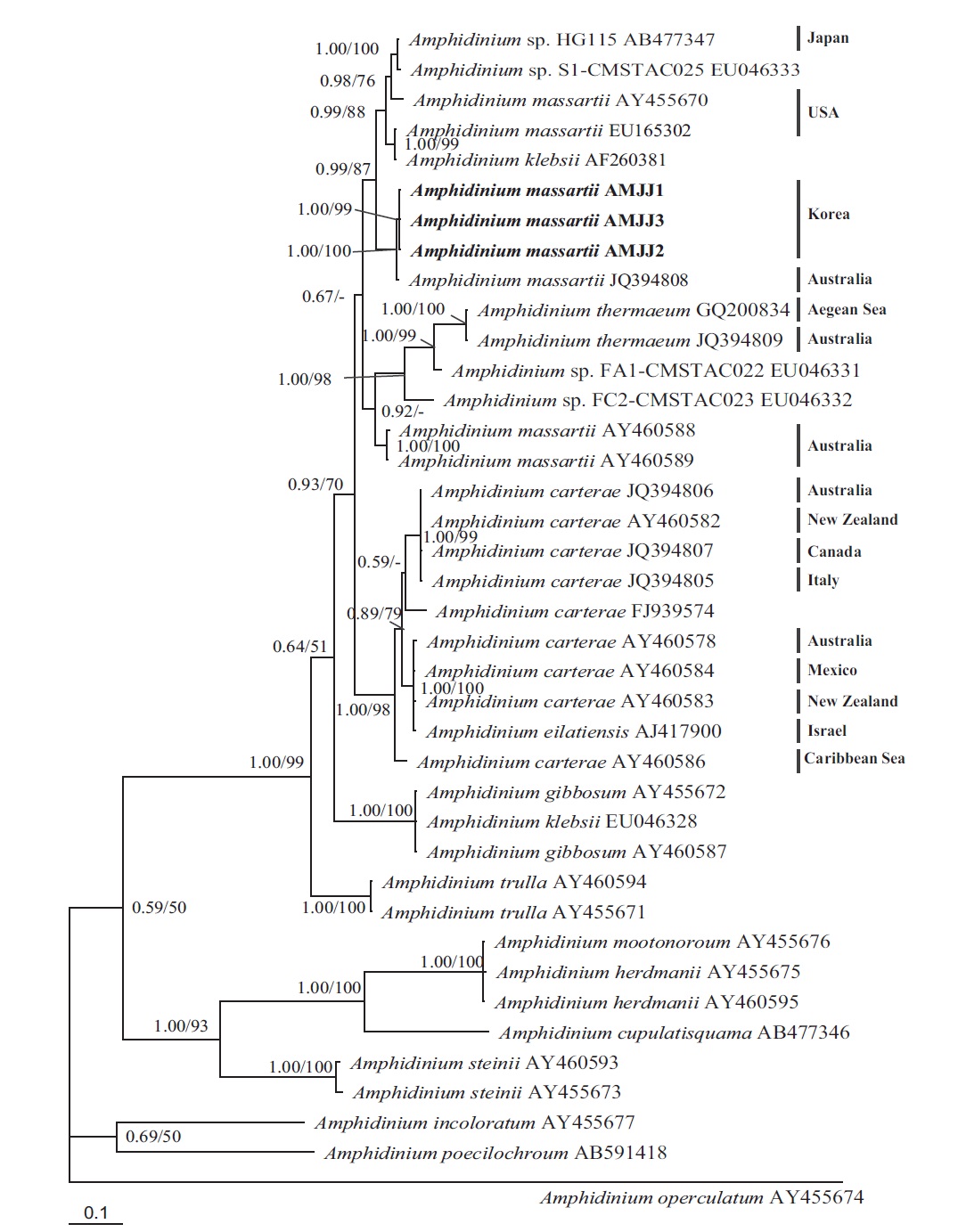
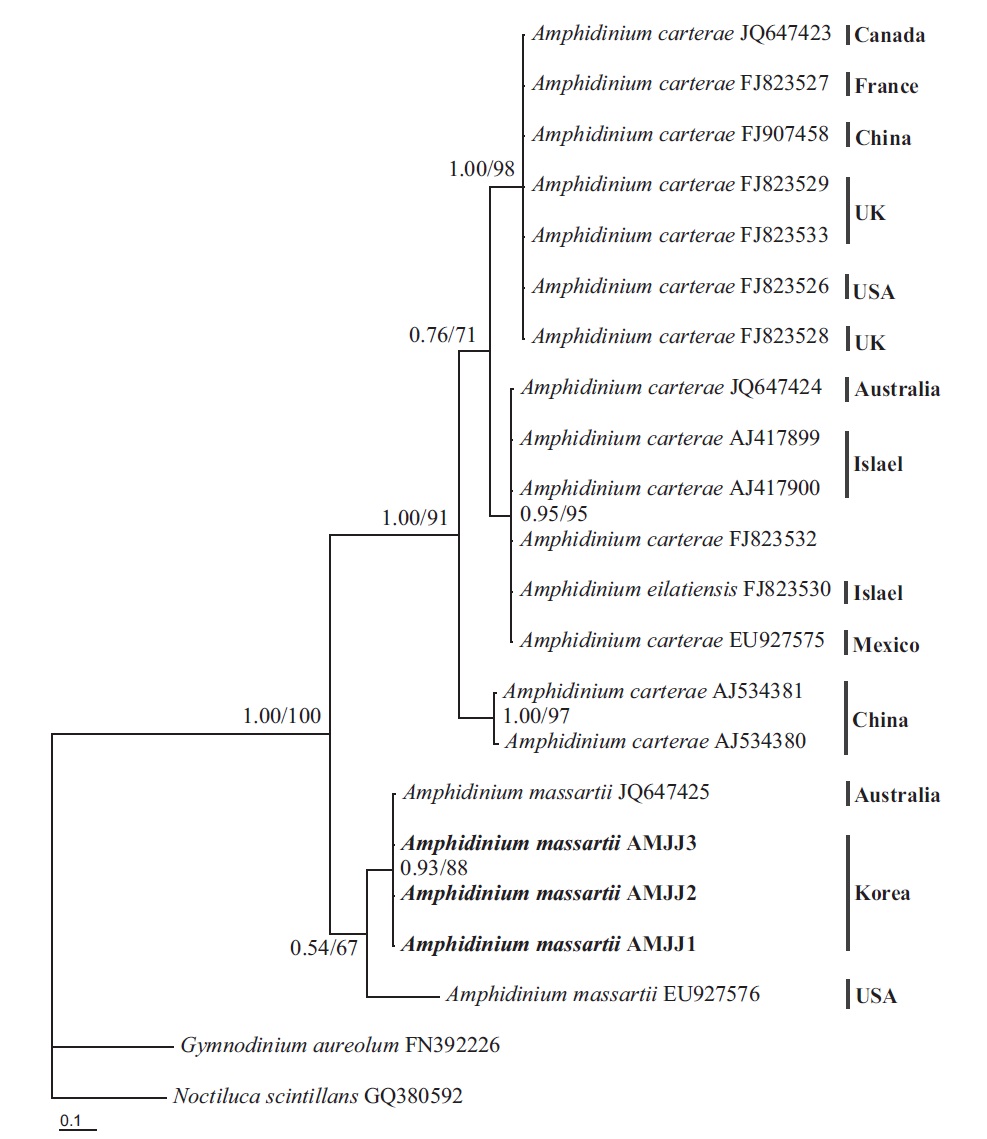
In the phylogenetic tree based on LSU rDNA sequences, with an Australian strain of A. massartii CS-259
(JQ394808), the three Korean strains of A. massartii formed a small clade that belonged to a larger clade consisting of A. massartii, Amphidinium thermaeum Dolapsakis and Economou, Amphidinium klebsii Kofoid and Swezy, and several unidentified Amphidinium spp. This large clade was divergent from another large clade consisting of A. carterae and Amphidinium eilatiensis J. J. Lee (Fig. 1). Furthermore, the small clade consisting of the Korean and Australian strains was also divergent from another small clade that consisted of the USA strains of A. massartii, the unidentified Okinawan strain of Amphidinium sp. HG115 (AB477347), and an unknown Amphidinium
sp. (EU046333).
In phylogenetic trees based on ITS rDNA sequences, the Korean strains of A. massartii formed a clade with the Australian strain of A. massartii and the USA strain of A. massartii was basal to this clade. In addition, the clade consisting of all A. massartii strains was clearly divergent from the large A. carterae clade (Fig. 2).
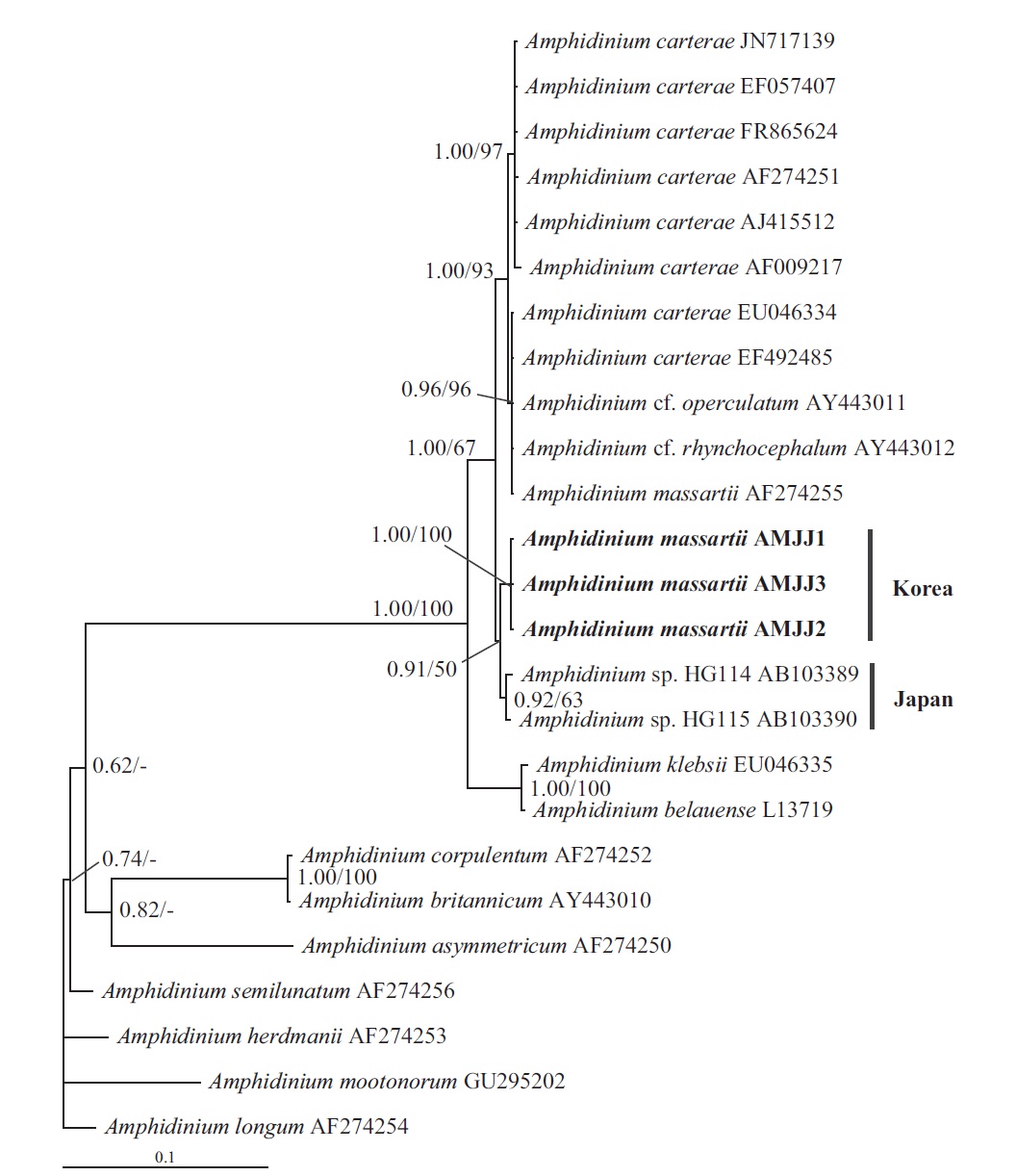
In this phylogenetic tree based on SSU rDNA, the Korean strains of A. massartii formed a small clade that was divergent from the Okinawan strains of Amphidinium sp. AB103389 (HG 114) and AB103390 (HG 115) (Fig. 3). However, while this divergence was somewhat weakly supported in ML, it was strongly supported by Bayesian analysis (0.91/50). A large clade containing these two
small clades was clearly divergent from a large clade consisting of all A. carterae strains, Amphidinium cf. rhynchocephalum, Amphidinium cf. operculatum, and a strain of A. massartii AF274255 (Fig. 3).
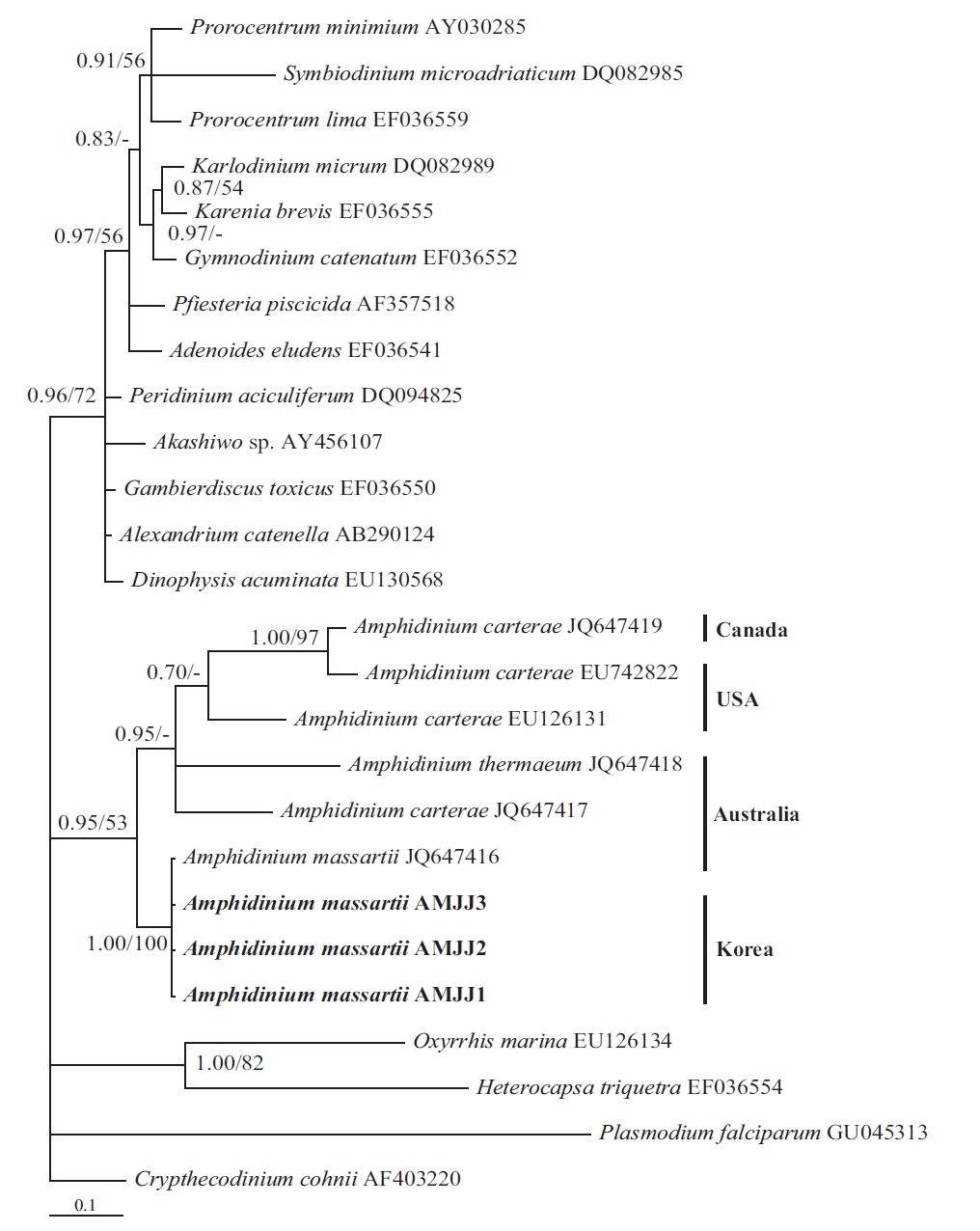
In the tree based on the cytochrome b sequences, the Korean strains and an Australian strain of A. massartii CS- 259 (JQ647416) formed a clade that was clearly divergent from the clade consisting of all A. carterae strains and A. thermaeum (Fig. 4).
On the basis of the sequences of rDNA and phylogenetic trees, the Korean strains of A. massartii have unique rDNA sequences, even though their sequences were similar to those of the Australian strain of A. massartii CS-259.
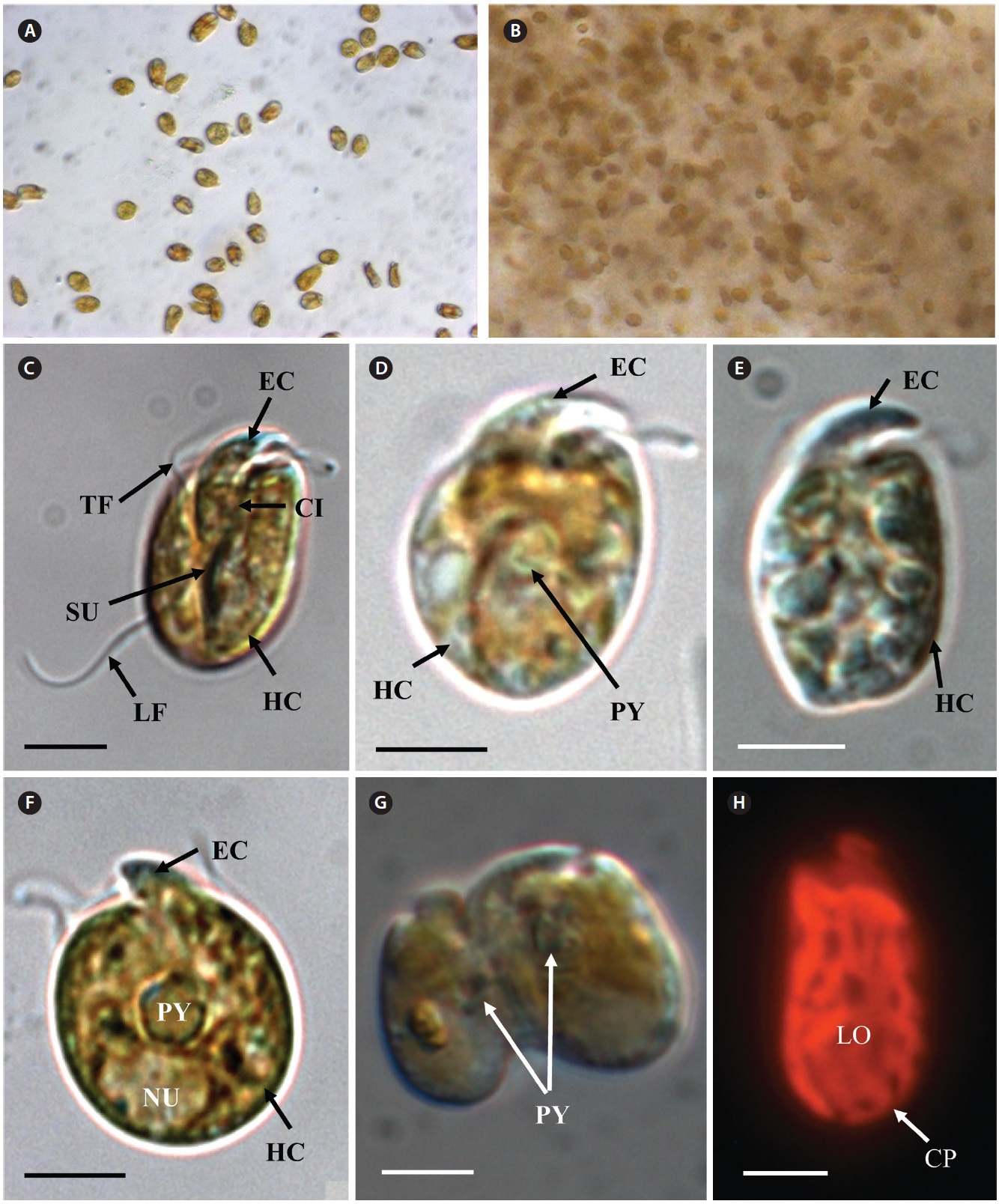
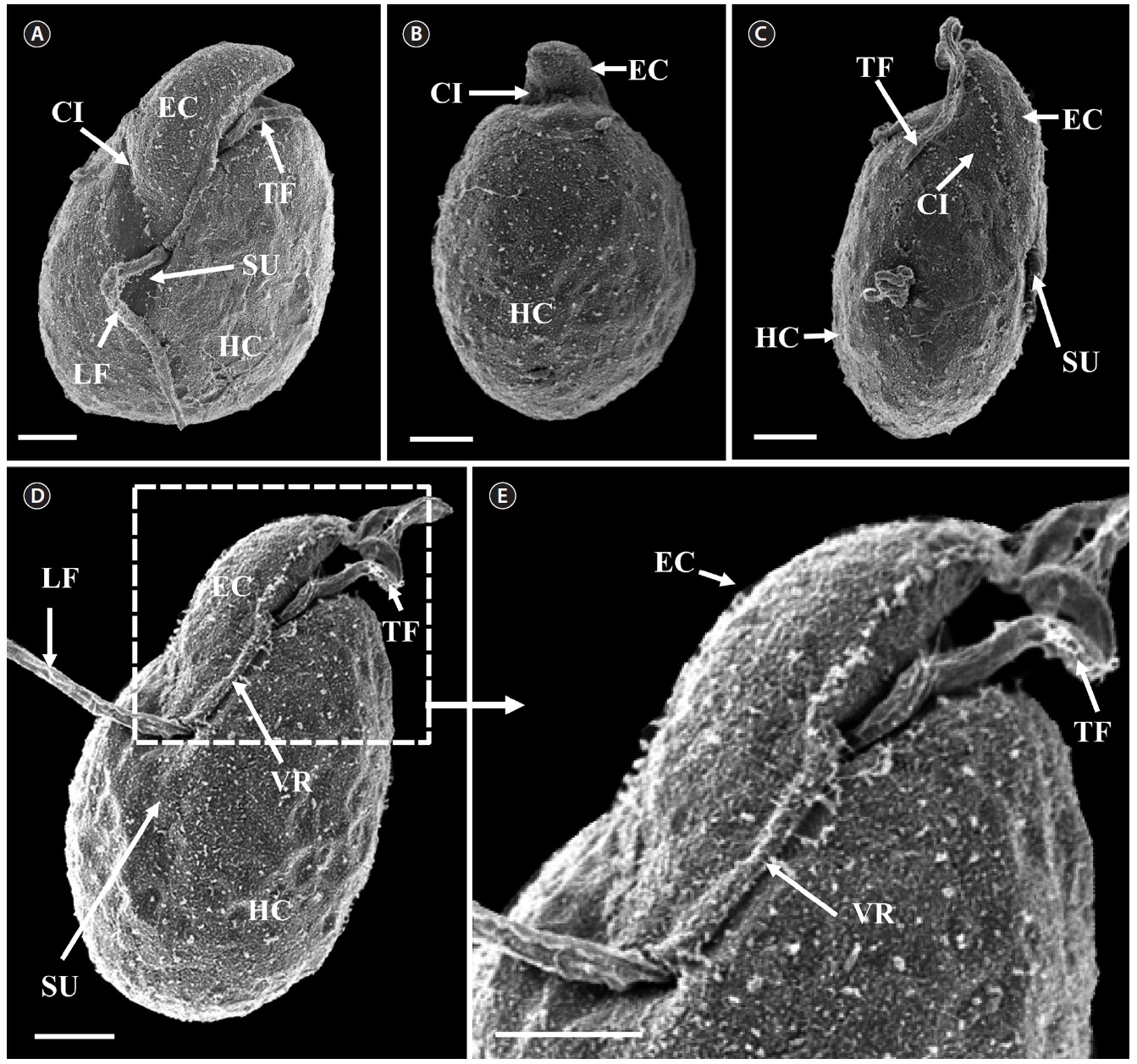
[Fig. 6.] Micrographs of the Korean strain of Amphidinium massartii AMJJ1 taken using scanning electron microscopy. (A) Ventral view showing the tongue-shaped epicone (EC), cingulum (CI), transverse flagellum (TF), sulcus (SU), longitudinal flagellum (LF), and hypocone (HC). (B) Dorsal view showing EC, CI, and HC. (C) Lateral view showing the dorso-ventrally flattened EC, TF, SU, CI, and HC. (D) Ventral view showing EC, LF, TF, SU, HC, and ventral ridge (VR). (E) Enlargement of Fig. 2D showing EC, TF, and VR. Scale bars represent: A-E, 2 μm.
In clonal cultures, most cells of the Korean strains of A. massartii swam freely, but some cells were embedded in mucilage (Fig. 5A & B).
Living cells were almost oval and dorso-ventrally flattened (Fig. 5). The epicone was tongue-shaped (Fig. 5C-F). The nucleus was round and positioned mainly in the posterior of the cell in hypocone (Fig. 5F). There was a single chloroplast that had globular lobes that radiated out from the central part of cell where the ring-like pyrenoid was observed (Fig. 5D, F & H). The length, width, and thickness of living cells were 12-17, 8-13, and 5-7 μm, respectively (Table 4).
Scanning electron microscopy indicated that the ratio
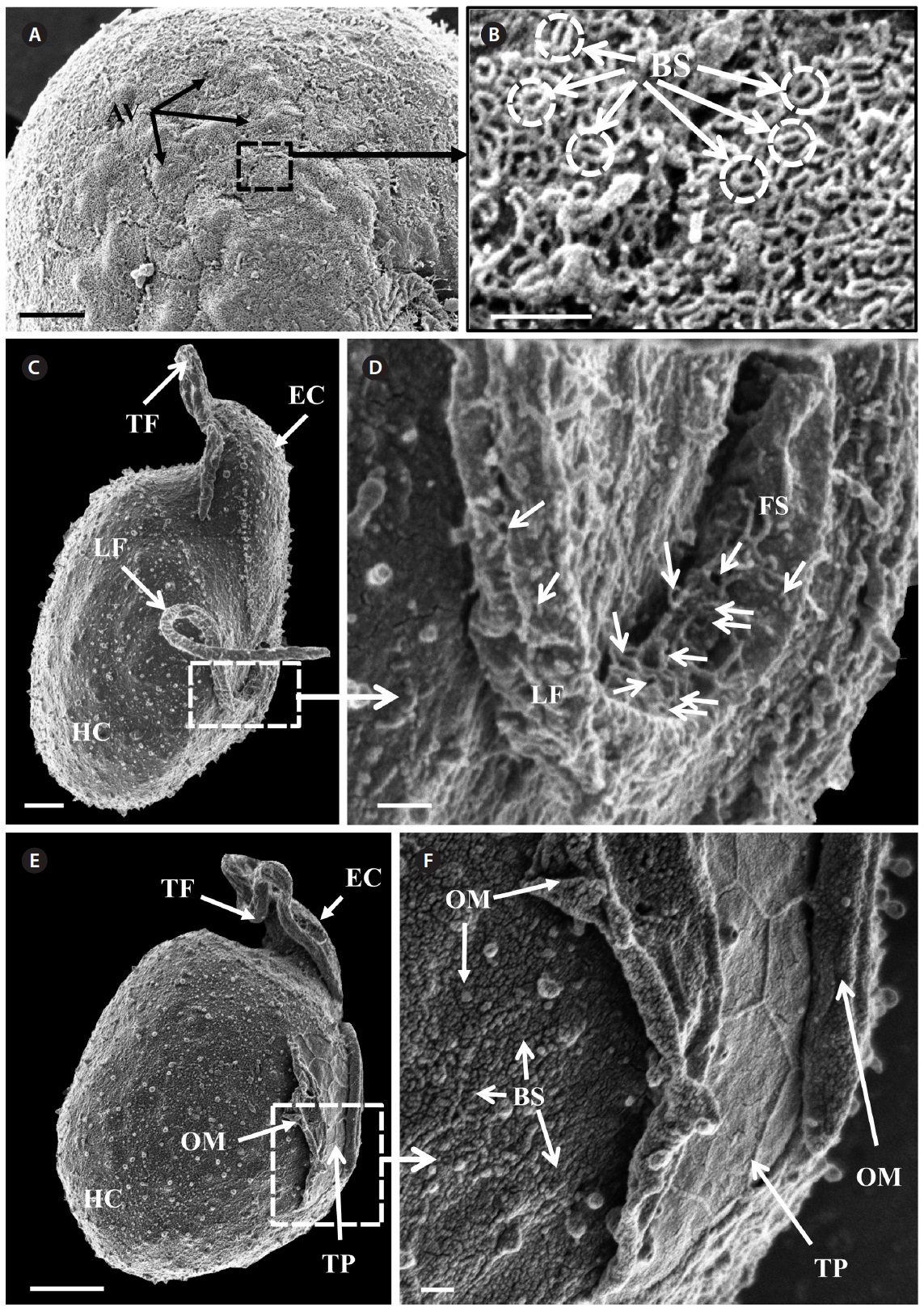
of the epicone relative to total cell length was approximately 1/2 (Table 4). The narrow cingulum was V-shaped and the narrow sulcus was also V-shaped (Fig. 6A & D). A ventral ridge was present and ran between the two points of flagellar insertion (Fig. 6D & E). The position of flagellar insertion was 0.4-0.5 of the cell body from the apex (Table 4). The amphiesmal vesicles were well observed in the outer membrane of some cells (Fig. 7A). In addition, a thin plate was observed (Fig. 7E & F). An oval-shaped, ring-like structure of body scales was observed in the outer membrane (Fig. 7B & F). In addition, comparablesized ring-like scales were observed on the flagella (Fig. 7D). Body scales were 50-70 nm in length and 40-50 nm in width (Table 4).
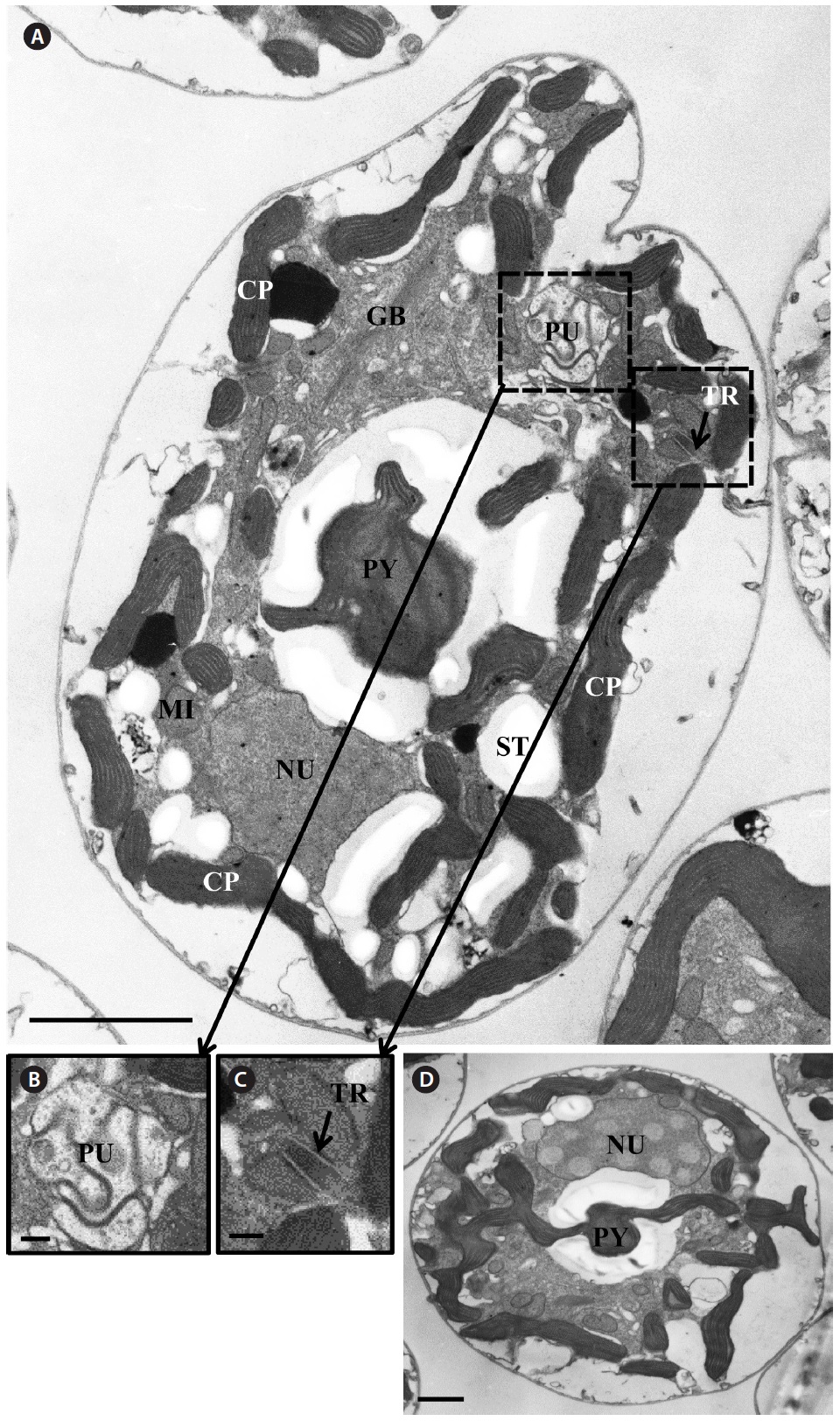
Transmission electron microscopy revealed the main ultrastructures, including a chloroplast, the nucleus, two stalked pyrenoids, starch grains, mitochondia, golgi bodies, a trichocyst, and a pusule (Fig. 8).
Cells of the Korean strain had chlorophyll a as the major pigment. In addition, cells had peridinin as the major carotenoid pigment and chlorophyll c2, chlorophyll a and diadinoxanthin as accessory pigments.
No nauplii of A. salina were dead after 48 h of incubation with A. massartii AMJJ1 concentrations of 100, 500, 1,000, 5,000, or 10,000 cells mL-1.
Phylogenetic trees based on ITS, and LSU rDNA and cytochrome b show that the Korean strains clearly belong to the Amphidinium massartii clade, which is clearly divergent from the A. carterae clade (Murray et al. 2012). This suggests that the Korean strains are A. massartii, not A. carterae. However, the Korean strains have unique rDNA sequences because there is no known Amphidinium species or strain with rDNA sequences that are identical to those of the Korean strains, even though the sequences of 5.8S and ITS2 rDNA and those of the mitochondrial cytochrome b gene of the Korean strains are identical to those of the Australian Queensland strain (Kurrimine Beach), A. massartii CS-259. The ITS1 and LSU sequences of the Korean strains are slightly (i.e., 0.6-0.7%) different from the Queensland strain. Considering the large distance
[Fig. 8.] Micrographs of the Korean strains of Amphidinium massartii AMJJ1 taken using transmission electron microscopy. (A) Longitudinal section showing several organelles inside the protoplasm. PY, pyrenoid; PU, pusule, TR, trichocyst; GB, Golgi body; CP, chloroplast; MI, mitochondrion; NU, nucleus; ST, starch. (B) Enlarged part of Fig. 8A showing PU. (C) Enlarged part of Fig. 8A showing TR. (D) Transverse section showing PY and NU. Scale bars represent: A, 2 μm; B & C, 200 nm; D, 1 μm.
between Jeju Island (Northwestern Pacific Ocean) and Queensland (Southwestern Pacific Ocean) (i.e., ~6,000 km), small differences in rDNA sequence and identical mitochondrial cytochrome b gene sequences imply that the Korean strains of A. massartii are closely related to the Queensland strain. The main current along Queensland is the East Australian Current (EAC), which is the western boundary current in the South Pacific and is similar to the Kuroshio Current that is the western boundary current in the North Pacific. The branched water mass of the Kuroshio Current passes Jeju Island (Ridgway and Godfrey 1997, Ichikawa and Beardsley 2002). The EAC is considered to be the South Pacific equivalent of the Kuroshio in the North Pacific (Ridgway and Godfrey 1997). Therefore, similar hydrography in the East Australian Current and the Kuroshio Current may cause the observed similarity in the genetics of the Queensland and Jeju Island strains of A. massartii. However, the sequences of rDNA of the Korean strains of A. massartii are considerably different from those of Tasmanian and USA (Rhode Island and Florida, North Atlantic Ocean) strains of A. massartii. Tasmania is located at a considerably higher latitude (41-43° S) than Kurrimine beach in North Queensland (17.8° S). The water temperature in North Queensland is 19-35℃, while that in Tasmania is 10-18℃ (Holdich and Harrison 1981, Coles et al. 1993, Hallegraeff et al. 1995). This difference in latitude and the associated differences in hydrography may result in the observed genetic difference between the strains of A. massartii found in Queensland and Tasmania. Exploration of this topic in greater detail would be worthwhile.
Murray et al. (2012) suggested that Amphidinium sp. HG114 and HG115 that were isolated from subtropical Okinawan waters may be A. massartii, even though their morphology has not been clearly confirmed to be that of A. massartii. The LSU rDNA sequences of the Korean strains were 4.5% different from those of Amphidinium sp. HG115, although the SSU rDNA sequences of the Korean strains were 0.9% different from those of Amphidinium sp. HG114 and HG115. In addition, in the tree based on the LSU rDNA sequences, the three Korean strains of A. massartii formed a small clade that was divergent from another small clade consisting of Amphidinium sp. HG115 and the USA strains of A. massartii. Therefore, the three Korean strains of A. massartii are genetically different from the Okinawan strains of Amphidinium sp.
The overall shape and characteristics of the Korean strains of A. massartii are similar those originally described for A. massartii (Biecheler 1952). Furthermore, the Korean strains have body scales and plastids that are sparse and consist of several lobes, as in Murray et al. (2012). We report, for the first time, the presence of scales on the surface of A. massartii flagella. Prior to the present study, only the heterotrophic dinoflagellate Oxyrrhis marina Dujardin was known to have both body and flagellar scales (Clarke and Pennick 1972, 1976). The phototrophic dinoflagellates Amphidinium sp. HG114, HG115, A. massartii CS-259, A. cupulatisquama, Heterocapsa spp., and Lepidodinium viride also have body scales, but scales on their flagella have not been reported (Pennick and Clarke 1977, Morrill and Loeblich 1981, Watanabe et al. 1990, Hansen 1995, Horiguchi 1995, 1997, Sekida et al. 2003, Iwataki et al. 2004, Tamura et al. 2009, Murray et al. 2012). Furthermore, Murray et al. (2012) suggested that A. carterae differs from A. massartii in that A. carterae does not have body scales. The results of the present study confirm this suggestion.
Zhang et al. (2007) suggested that in the phylogenetic tree based on dinoflagellate cytochrome b, the genera Oxyrrhis, Heterocapsa, and Amphidinium, which possess body scales, are placed in the basal position. Therefore, the possession of body scales may be an important feature in dinoflagellate evolution and it will be worthwhile exploring this topic in detail.
For the Korean strains of A. massartii, the position of flagellar insertion is 0.4-0.5 of the cell body from the apex. Murray et al. (2012) suggested that flagellar insertion of A. massartii (~0.6 of the cell body from the apex) is situated in a lower position than that of A. carterae (~0.4). Therefore, differences in the position of flagellar insertion may be a critical key for differentiating A. massartii from A. carterae. Further examination of other strains is necessary in order to clarify this key.
Prior to the present study, A. massartii had been reported in Seto, France; British Columbia, Canada; Rhode Island and Florida, USA; and Queensland and Tasmania, Australia (Biecheler 1952, Baillie 1971, Jørgensen et al. 2004, Murray et al. 2004, 2012), but both DNA sequences and morphology have been reported for only the Australian and USA strains (Fig. 9). Therefore, information about both the DNA sequences and morphology of the Korean strains of A. massartii in this study may contribute to resolving some of the ambiguity in the taxonomy of A. massartii and A. carterae. The discovery of the presence of A. massartii in the coastal waters of Jeju Island extends the global distribution of this species to the temperate region of the northwestern Pacific (33.3° N).
The results of the present study suggest that the Korean strains of A. massartii are not toxic. Murrary et al. (2011) reported that the Queensland strain of A. massartii CS- 259 was also non-toxic. Many strains of A. carterae are known to be toxic (Nayak et al. 1997, Jeong et al. 2001, Baig et al. 2006). Therefore, it will be worthwhile investigating the toxicity of other stains of A. massartii to determine whether non-toxicity can be used as a key for differentiating A. massartii from A. carterae.