Ginseng has been most widely used as herbal medicine in Eastern Asian countries, including Korea, China, and Japan. Ginseng is a species of a perennial plant belonging to the genus
Lung cancer, comprising of a majority of non-small-cell lung carcinomas (NSCLCs) of up to 85%, is the second leading cause of cancer death worldwide [9]. Thus, lung cancer is still considered as an extremely lethal cancer [10]. Surgical and combined therapies have contributed to reducing the mortality and the morbidity of lung cancer over last several decades; however, these therapies have limitations. Thus, complementary and alternative therapies with herbal medicine have recently been introduced in the Western medical society. However, few studies have reported on the anti-cancer activity of regular ginseng extract on a molecular biological level.
In this study, we prepared a modified regular ginseng extract (MRGX) that reinforced some gensenosides from regular ginseng butanol-extract (GBX) by using treatments with enzymes such as laminarinase and pectinase for stronger anti-cancer effect. The assessments of the gene expression pro?les in human lung cancer samples by using cDNA microarray technology showed that some genes were down-regulated while others were up-regulated during the MRGX-treatment process. Because gene expression might change during the lung cancer cell death process, such studies should use speci?c components and not serum that contains a mixture of gensenosides.
2.1. Preparation of MRGX extract
The root of regular ginseng (4 yr old) was purchased from the National Agricultural Cooperative Foundation in Chuncheongnam-do, Korea. A total of 20 g of pulverized ginseng root powder was suspended in 380 ml of distilled water and then sterilized at 121℃ for 15 min. The ginseng butanol extract (GBX) contained compounds with specific anti-lung cancer activity. We used GBX as a control of regular ginseng. In addition, the suspension was treated with an aliquot of filter-sterilized commercial enzymes (laminarinase: 100 mg, pectinase: 100 mg) with equimolar ratio (1:1, specific activity unit); the mixture was then incubated at 40℃ for 2 days and evaporated to dryness at 60℃. These enzymes-modified ginseng powders were suspended in 400 ml of 80% (v/v) methanol. The suspension was treated in an ultrasound bath for 5 min and filtered through Whatman No. 2 filter paper. The filtrates were combined and evaporated to dryness at 50℃. The extract was diluted to a concentration of 10% (w/v) in 70% ethanol. In the present study, the finally-obtained sample was called the modified regular ginseng extract (MRGX).
Human lung cancer (A549) cells were obtained from American Type Culture Collection (Rockville, MD). Cell lines were grown in DMEM (Dulbecco Modified Eagle’s Medium, Sigma, USA) supplemented with 10% (v/v) FBS (Fetal Bovine Serum, GIBCO, NY) and 1% (w/v) penicillin-streptomycin (GIBCO, NY). The cells were incubated at 37℃ with 5% (v/v) CO2 for 24 h. The density of A549 cells was adjusted to 5 × 103 cells/well in a 96-well plate. After a 24-h incubation, the cells were treated with MRGX at 50 ug/ml concentrations. The appropriate dose was determined by evaluating the cytotoxicity of MRGX for 24 h. To the cell solution, 10 ul of cell-counting kit-8 solution (Dojindo, Japan) was added for 1 h. Cell viability was determined by using a microplate reader (Sunrise, Tecan, Switzerland) to measuring the absorbance at 450 nm.
For the microarray analysis of the MRGX-treated lung cancer cells, a human twin 44K cDNA chip was used for the transcription profiling analysis. Total RNA was extracted from vehicle- or MRGX-treated lung cancer cells, and cDNA probes were prepared by using reverse transcription of 50 mg of RNA in the presence of aminoallyl-dUTP, followed by coupling with Cy3 dye (vehicle) or Cy5 dye (MRGX-treated). The mixed Cy3- and Cy5-labeled RNA from lung cancer cells (A549) was hybridized with one side of the Twin ChipTM Human 44K (Genocheck, Seoul, Korea), and that from MRGX-treated cells was hybridized with the other side of the chip.
Fluorescent images were quanti?ed and normalized, as described previously [11]. This experiment was repeated four times. Genes were considered to be differentially expressed when the global M value, log2 (R/G), exceeded |1.0| (twofold) with a
2.4. Ontology-related network analysis
To study the biological functions of ontology-relate regulated genes and proteins through their interaction network, we conducted a bioinformatic protein network analysis by using an ingenuity pathways analysis (IPA, http://www.ingenuity.com). The IPA identifies the protein interaction network on the basis of a regularly-updated “Ingenuity Pathways Knowledge-base.” The updatable database containing millions of individual protein-protein relationships was retrieved from the biological literature. Network generation was optimized to include as many proteins from the inputted expression profile as possible and aimed at the production of highly-connected networks.
2.5. Semi-quantitative RT-PCR confirmation
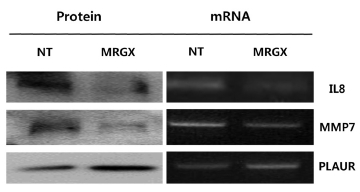
Total RNA was extracted from young and senescent cells by using an acid guanidinium thiocyanate phenol-chloroform extraction-based method. To compare the relative amounts of mRNA in young and senescent cells, we performed a semi-quantitative RT-PCR. A series of mixtures was prepared by mixing RNA from young and senescent cells as indicated, so that each mixture had the same total amount of RNA (2 μg) in a constant volume (12 μl). The RT reaction was carried out in a final volume of 20 μl by using Superscript II reverse transcriptase according to the manufacturer’s protocol, and 4 μl of the final RT product mix was then PCR amplified. The primer sets used were 5’-ATCTGGCAACCCTAGTCTGC-3’ and 5’-GTGCTTCCACATGTCCTCAC-3’ for IL8, 5’-GACATCATGATTGGCTTTGC-3’ and 5’-TCCTCATCGAAGTGAGCATC-3’ for MMP7 and 5’-GTGAGGAAGCCCAAGCTACT-3’ and 5’-ATGTCCAAGGTGGCTTCTTC-3’ for PLAUR.
Expression levels of signaling proteins were examined by using Western blot analyses. In brief, 30 μg of denatured protein were run using 12% polyacrylamide gel electrophoresis and then transferred onto a nitrocellulose membrane. The transferred nitrocellulose membrane was stained with Ponceau S to position the proteins. The blotted membrane was blocked for 1 h with 5% (w/v) skimmed milk in TTBS (Tween-20 and Tris-buffered saline), followed by incubation with a dilution of primary antibodies (Cell Signal, Boston, USA), including interleukin-8 (IL-8), matrix metallopeptidase 7 (MMP7) and plasminogen activator, urokinase receptor (PLAUR), at room temperature for 2 h or at 4℃ overnight. The membrane was washed three times for 5 min with 0.1% (v/v) TTBS before incubation with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG or HRP-conjugated rabbit anti-goat IgG with a 1:2000 dilution in TBS containing 5% (w/v) skimmed milk at room temperature for 1 h. The membrane was rinsed three times with TBS-0.1% (v/v) Tween-20 for 5 min. The Pierce® ECL system (Thermo Scientific, San Jose, CA) was used to develop the proteins on an X-ray film (Kodak, Seoul, Korea). The expression levels of proteins were quantitatively analyzed with GAPDH as an internal control.
3.1. Analysis of MRGX related gene expression in lung cancer cells
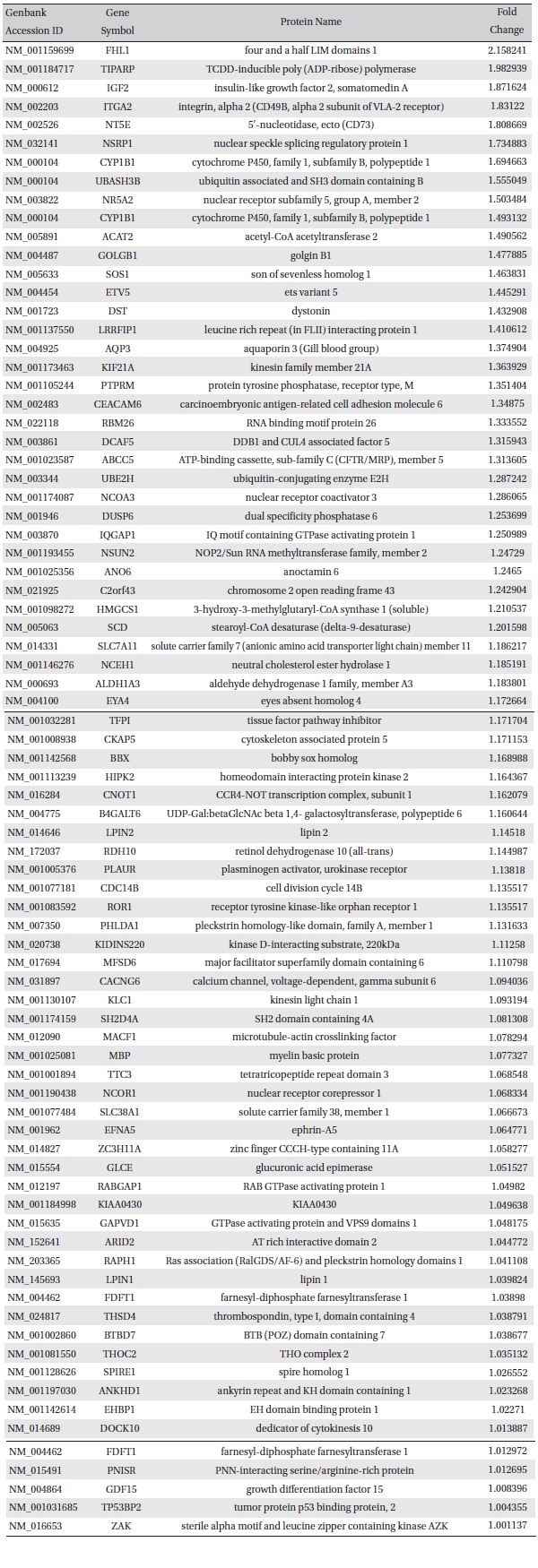
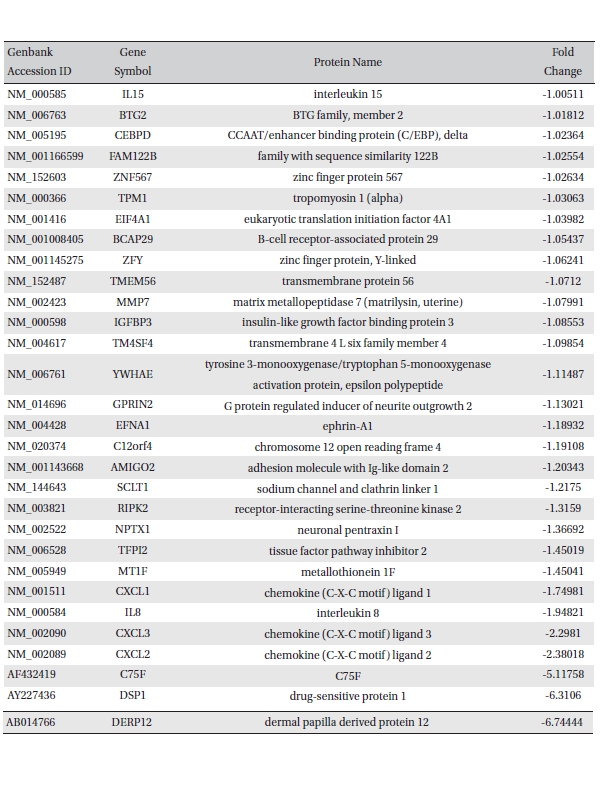
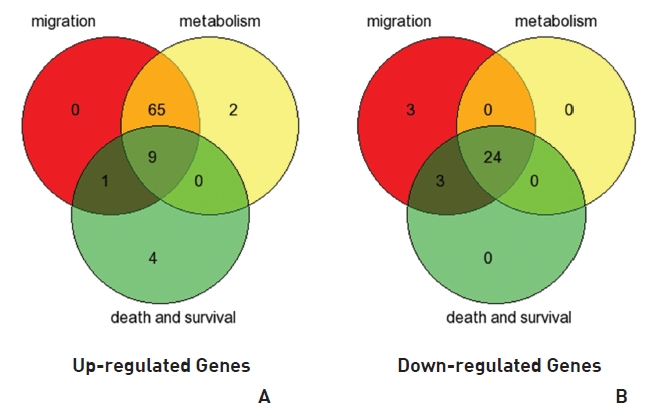
To analyze the MRGX-related gene expression in lung cancer cells, we used a cDNA microarray analysis approach. Microarray data were filtered and combined using gene symbols; then, network analyses were performed. The clustered microarray data showed that groups of genes in lung cancer and MRGX-treated lung cancer cells were regulated differently by 50 mM of MRGX. Genes with at least one global M |1.0| are shown in (Table 1) and 2. A total of 109 were ultimately identified and categorized in terms of their cellular functions by using gene ontology and IPA annotation: The Venn-diagram for migration (45%), metabolism (45%), and cell death and survival (10%) are shown in (Fig 1) The relative abundances of the regulated genes between lung and MRGX-treated lung cancer cells determined by using a semi-quantitative analysis, as described previously [12], are listed in (Table 1) Based on the criterion of a fold change > 1.0, 80 were up-regulated (Fig. 1A) whereas 29 were down-regulated in MRGX-treated lung cancer cells (Fig 1)B) compared to lung cancer.
3.2. Network analysis based on Gene Ontology analysis
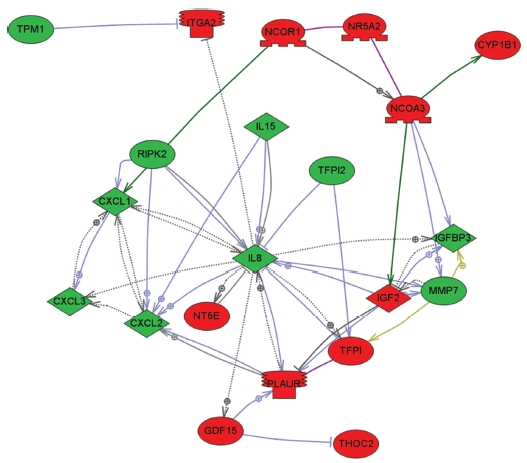
To explore key MRGX-related proteins in the gene ontology analysis of gene functions, we used IPA to query 109 proteins belonging to the proteins up- and down-regulated by MRGX, resulting in a distinct interconnected network of 21 proteins (Fig 2) There were quantitative and qualitative alterations in MRGX-treated lung cancer cells between the regulation groups. Among them, interleukin 8 (IL8) was identified as the center of the MRGX-related protein network in lung cancer cells. IL-8 is a lung giant-cell carcinoma-derived chemotactic protein that binds CXCR1 and CXCR2 [13]. In the present study, IL-8 is one of critical chemo-attractants responsible for leukocyte recruitment, cancer proliferation, and angiogenesis, and the potential mechanism of IL-8 production from human non-small-cell lung cancer was investigated [14]. IL-8 was decreased dramatically in the MRGX-treated lung cancer cells. The protein-protein network analysis suggests that IL-8 is a major protein that interacts with multiple proteins and that it is directly or indirectly down-regulated or up-regulated in MRGX-treat lung cancer cells. IL-8-linked proteins include chemokine (C-X-C motif) ligand 2 (CXCl2), MMP7 and PLAUR. PLAUR plays a key role in lung cancer [15]. However, the detailed mechanism has not been studied in lung cancer. Herein, the level of IL-8 in the MRGX-treated lung-cancer cells was 1.9-fold down-regulated while PLAUR was up-regulated. The accumulation of PLAUR has been associated with lung cancer therapy.
[Table. 1] Up-regulated gene list
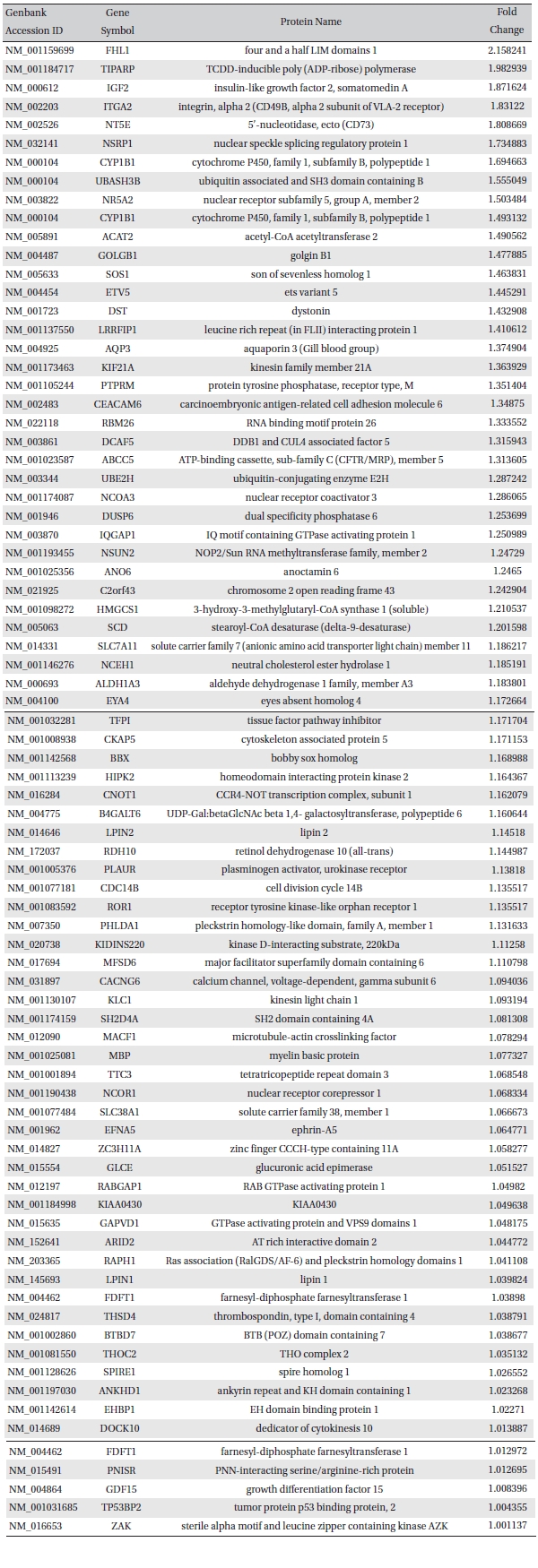
Up-regulated gene list
[Table. 2] Down-regulated gene list
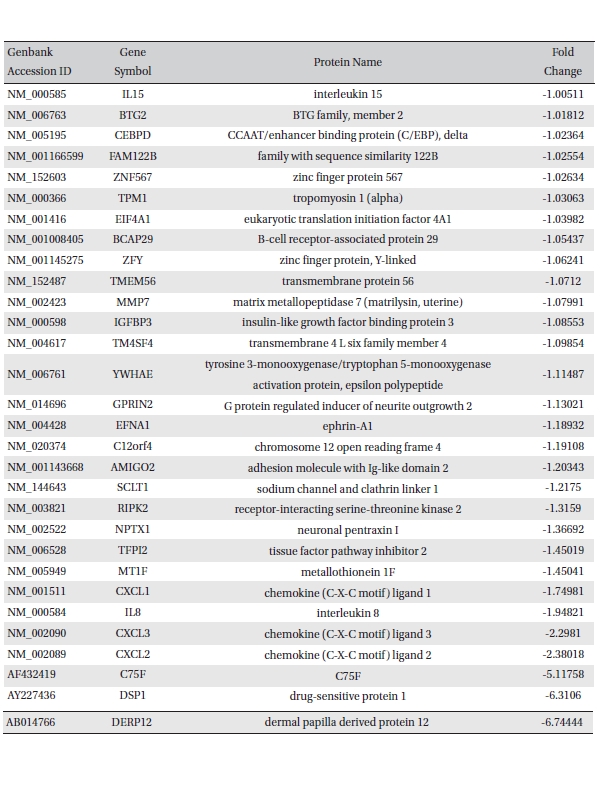
Down-regulated gene list
Microarray analyses may result in biased data, and inconsistent results are often obtained when different methods are used. Therefore, the data need to be validated with a specific method. Because we measured gene expression, we wanted to validate our screening results with conventional methods such as RT-PCR and Western blots. We selected genes from each of the two up- and down-regulated group of genes for validation by using semi-quantitative RT-PCR and Western blot analyses. Four genes were selected based on their high fold-change (a large number) in the analysis and the availability of commercial antibodies. IL-8, MMP7and PLAUR are examples of up- and down-regulated genes. The mRNA levels of IL-8 and MMP7 were decreased in the MRGX-treated lung cancer cells, and the protein levels of IL-8 and MMP7 also decreased. The semi-quantitative PCR and Western blot results showed that PLAUR was increased (Fig 3)
In conclusion, most MRGX-responsive genes are up-regulated transiently in A549 cells, but are down-regulated in a sustained manner in lung cancer cells. These genes might be involved in the altered MRGX responsiveness observed during the cancer migration and metabolism processes. The roles of these genes in MRGX responses, including the cell cycle and cell growth, should be evaluated in further studies by modulating their expressions.