During insect development the central nervous system regulates growth, metamorphosis, diapause and reproduction by secreting peptides. Insect hemolymph contains various proteinaceous components viz., transport proteins, storage proteins, peptide hormones which are essential for growth and development, and defensive peptides while induced by bacterial invasion or injury. Injection of the hemolymph obtained from larvae of other
This ENF peptide family shares similar amino acid sequence and conserved at C-terminal region. It is named after the N-terminal-conserved three amino acids (Glu-Asn-Phe) (Strand
In
In
There are several 30KPs as well as their homologs reported in silkworm
In a recent study by Zhang
The abundant level of 30KPs expression suggests that they may serve as storage proteins (Fujiwara and Yamashita, 1992; Izumi
In this study we report a high expression of the paralytic peptide and paralytic peptide binding protein in the diapause induced eggs of multivoltine silkworm
The multivoltine strain MW13 (Indian origin) was selected for the study. The larvae were reared as per the standard rearing method of Krishnaswami., (1978) up to last instar to obtain non-diapausing eggs, The late stage (4th & 5th instars) larvae were then reared under low temperature (18℃) and photoperiod (6L: 18D) up to cocooning stage and the moths were made to lay diapausing eggs at normal room temperature (25℃) (Saravanakumar
After oviposition, the diapause and non-diapause egg samples were collected from 0 to 48h at every 6h time interval, while, other tissues were collected from the day 3 of 5th instar larvae. Total RNA was extracted from the diapause and non-diapause eggs using TRIzol reagent (Invitrogen, USA), denatured in formaldehyde, formamide and electrophoresed in 2.0% agarose gels.
The first strand cDNA was synthesized utilizing RNA (2 μg) treated with 0.5μl of DNase buffer and 0.5 μl of DNase (Invitrogen, USA) for 15 minutes. Then, the reaction was terminated by heating at 75℃ for 10 minutes, to the above DNase treated sample, 1μl 10 mM dNTP, 1μl oligo (dT)18 (0.01mM) (Eurofin India Pvt Ltd, Bangalore) was added followed by incubation at 65℃ for 5 min. Finally 1X reverse transcriptase buffer (4 μl), 5 mM DTT (1 μl) and 1μl of M-MLV Superscript III reverse transcriptase (Invitrogen, USA) was added to obtain a final volume of 20 μl. The reaction was terminated by heating at 75℃ for 10 min according to the manufacturer’s protocol.
>
Microarray experiment and data analysis
A genome wide oligonucleotide microarray containing 24,924 probes were used to investigate the gene expression profiles of diapause induced and nondiapause eggs of multivoltine silkworm
PCR amplification was performed in a 25 μl reaction mixture containing 2.0 μl of 10X reaction buffer (100 mM Tris-HCL, pH 8.3, 500 mM KCl), 0.2 mM dNTPs, 1.5 mM MgCl2, 10 picomoles of forward and reverse primers, 0.3 U of Taq DNA polymerase (MBI Fermentas) with 1μl first strand cDNA as template. β actin (FP 5’cactgaggctcccctgaac 3’ and RP 5’ ggagtgcgtatccctcgtag 3’) (Eurofins, Bangalore) was used as an internal standard. The PCR amplification was carried out under the following conditions: 94℃ for 3min followed by 27 cycles of 94℃ for 30s, 54℃ for 30s, 72℃ for 2 min and a final extension of 7 min at 72℃.
One μl of first strand cDNA synthesized from diapause and non-diapause eggs from 6 to 48h after oviposition was used as template for qPCR analysis in a 25μl reaction mixture containing SYBR green mastermix
(ABI, CA, USA) and the specific primers designed for Realtime PCR (qPCR). The reactions were conducted on a STRATAGENE Mx 3005P realtime PCR system. The experiment was performed in triplicate and results were standardized to the expression level of the constitutive β actin gene. A Non-template control (NTC) sample was also run to detect contamination if any.
>
Identification of paralogous gene sequences in B.mori
The
Oligonucleotide microarrays containing 24,924 probes were used to investigate the gene expression profiles of diapause induced and non-diapause eggs of multivoltine silkworm B.mori at 18 and 30hrs after oviposition. The complete sets of raw and normalized data from this study have been deposited in the NCBI Gene Expression Omnibus (GEO) repository (accession number GSE35622).
A total of 638 genes were upregulated and 1136 genes were down regulated at 18 h after oviposition, whereas, 675 genes were found to be upregulated and 595 genes down regulated at 30 h after oviposition. Further, genes whose expression was detected in diapause induced and non-diapause silkworm eggs were classified into two groups, stably expressed and variably expressed genes at both 18 and 30 h. 115 genes were stably upregulated, while 117 genes stably down regulated at both 18 as well as 30 h. Up and down regulated genes showing fold change of 0.8 and
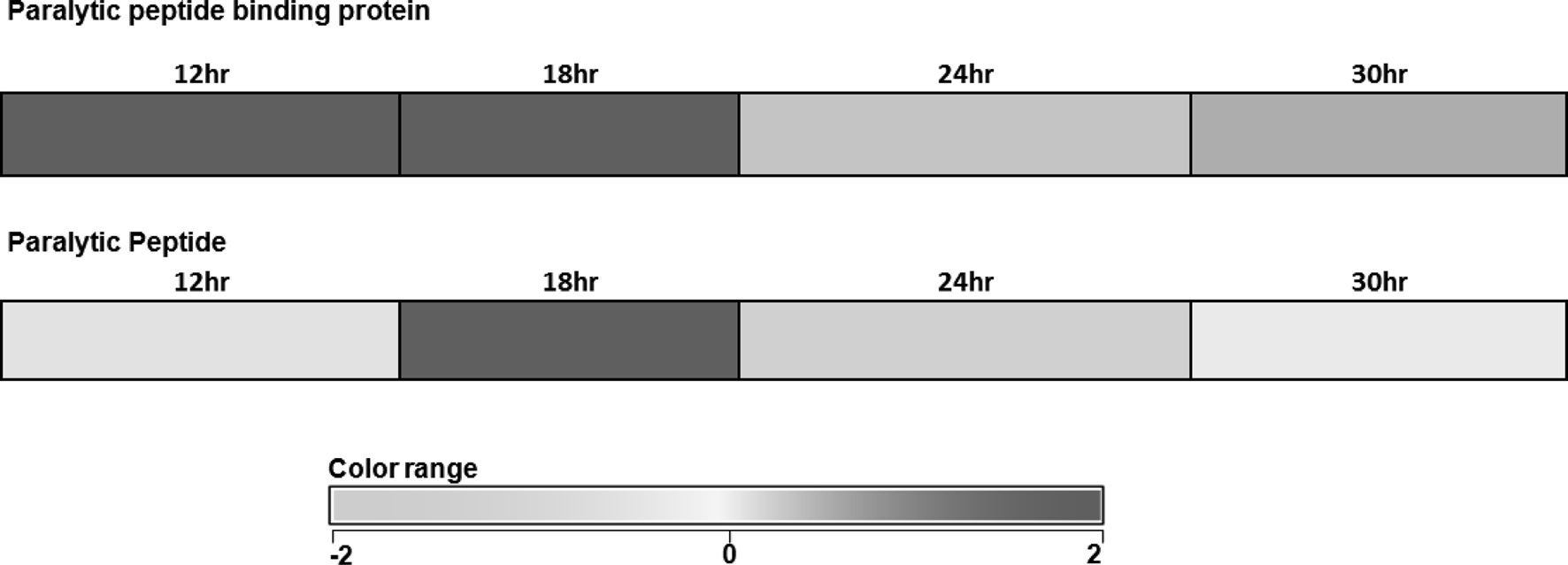
above were considered as significantly expressed. 315 genes were found to be variably upregulated at 18 h and 259 genes were found to be variably up regulated during 30 h of diapause-induced eggs (Unpublished data). Among those upregulated genes PP and PPBP were significantly up-regulated at 12 and 18 hours (Fig. 1). Further these two gene expressions were analyzed in diapause induced eggs at different time intervals after oviposition.
>
PP-BP gene expression in different tissues
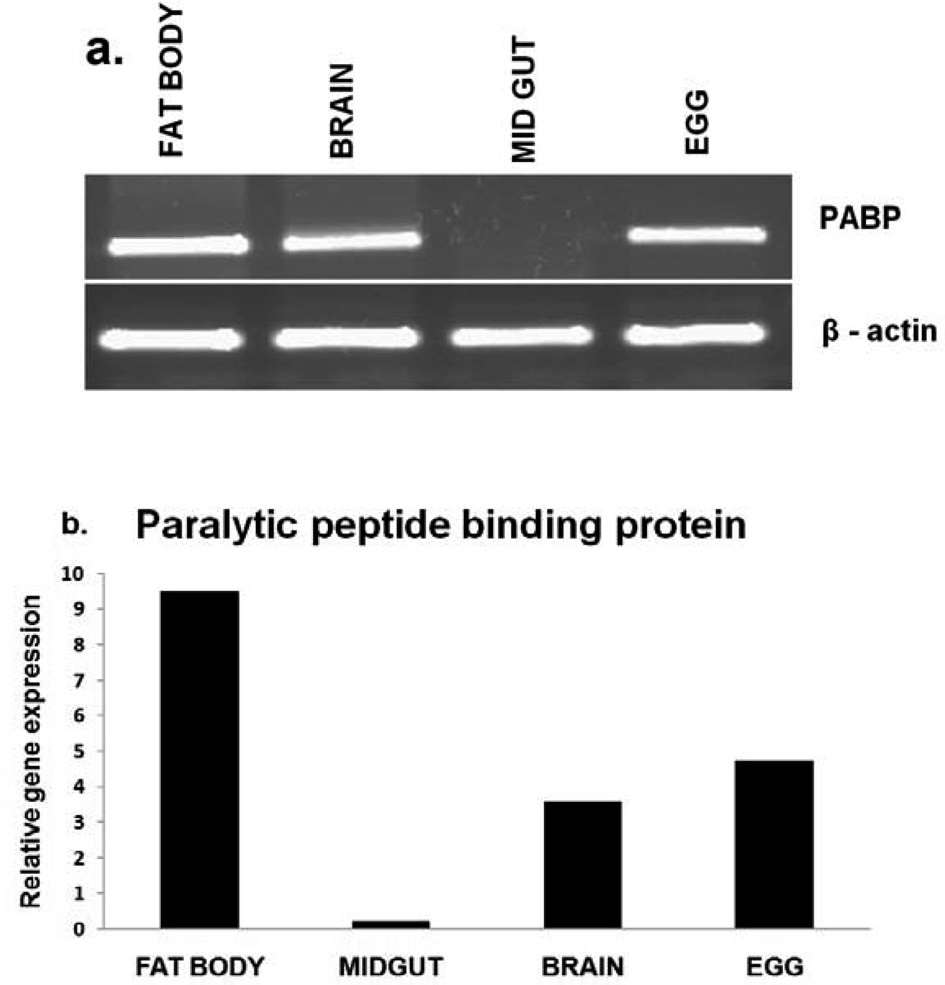
RT-PCR analysis was performed to determine the tissue specific expression of the
control. Further the results validated through realtime PCR analysis (Fig. 2. b) revealed that the PP-BP gene expression was higher in diapause induced eggs, compared to the non-diapause eggs.
>
Differential expression of B. mori PP-BP and PP in diapause induced eggs
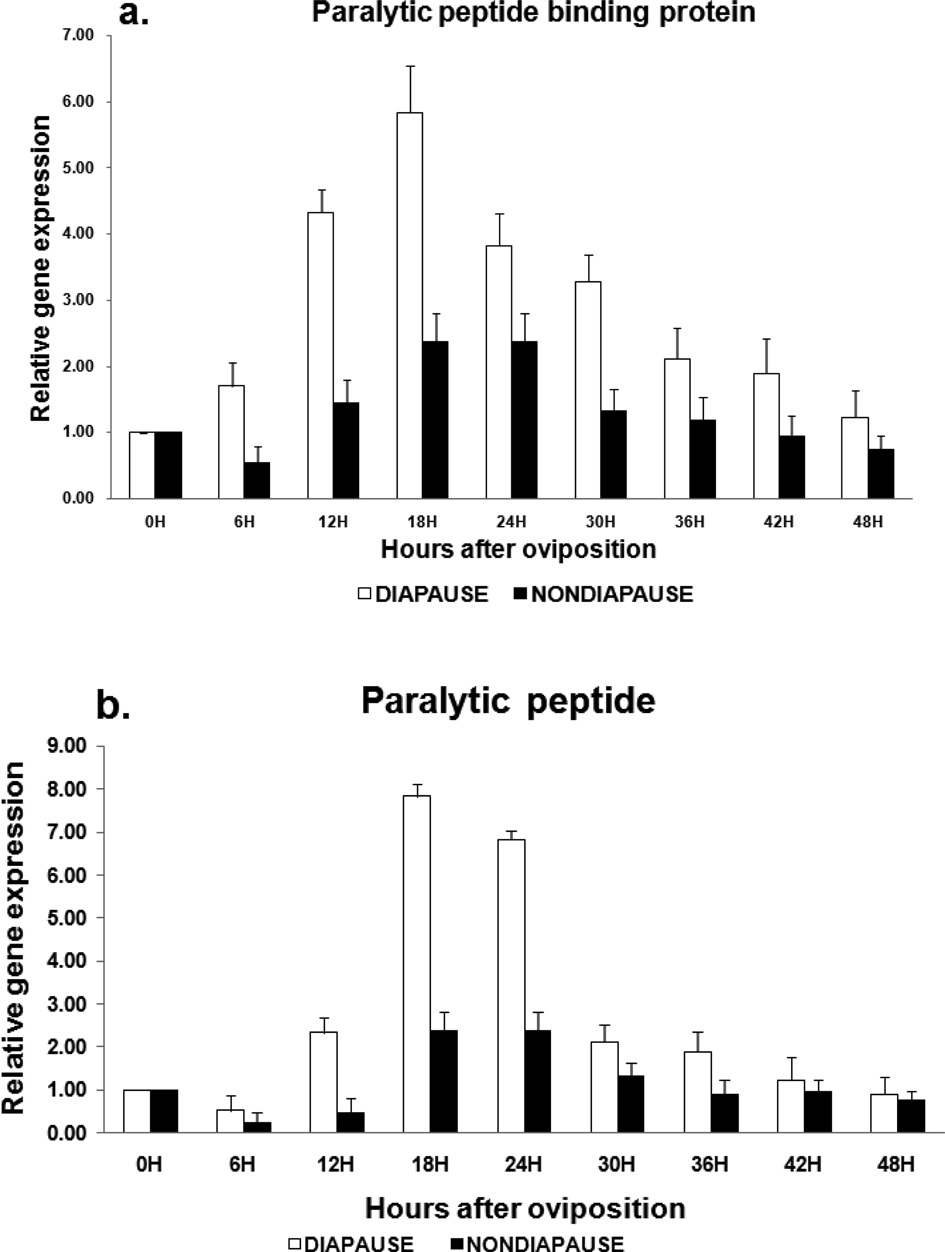
The expression level of PP-BP gradually increased from 0 to 24 h, where, the expression was at maximum, followed by a very steep decrease from 30 to 48 h in diapause induced eggs. The gene expression in non-diapause eggs was very low from 6 to 48 h time intervals compared to diapause eggs. The expression level of paralytic peptide was almost similar to PP-BP in diapause eggs while in non-diapause eggs they were slightly higher compared to PP-BP. (Fig. 3a, 3b).
>
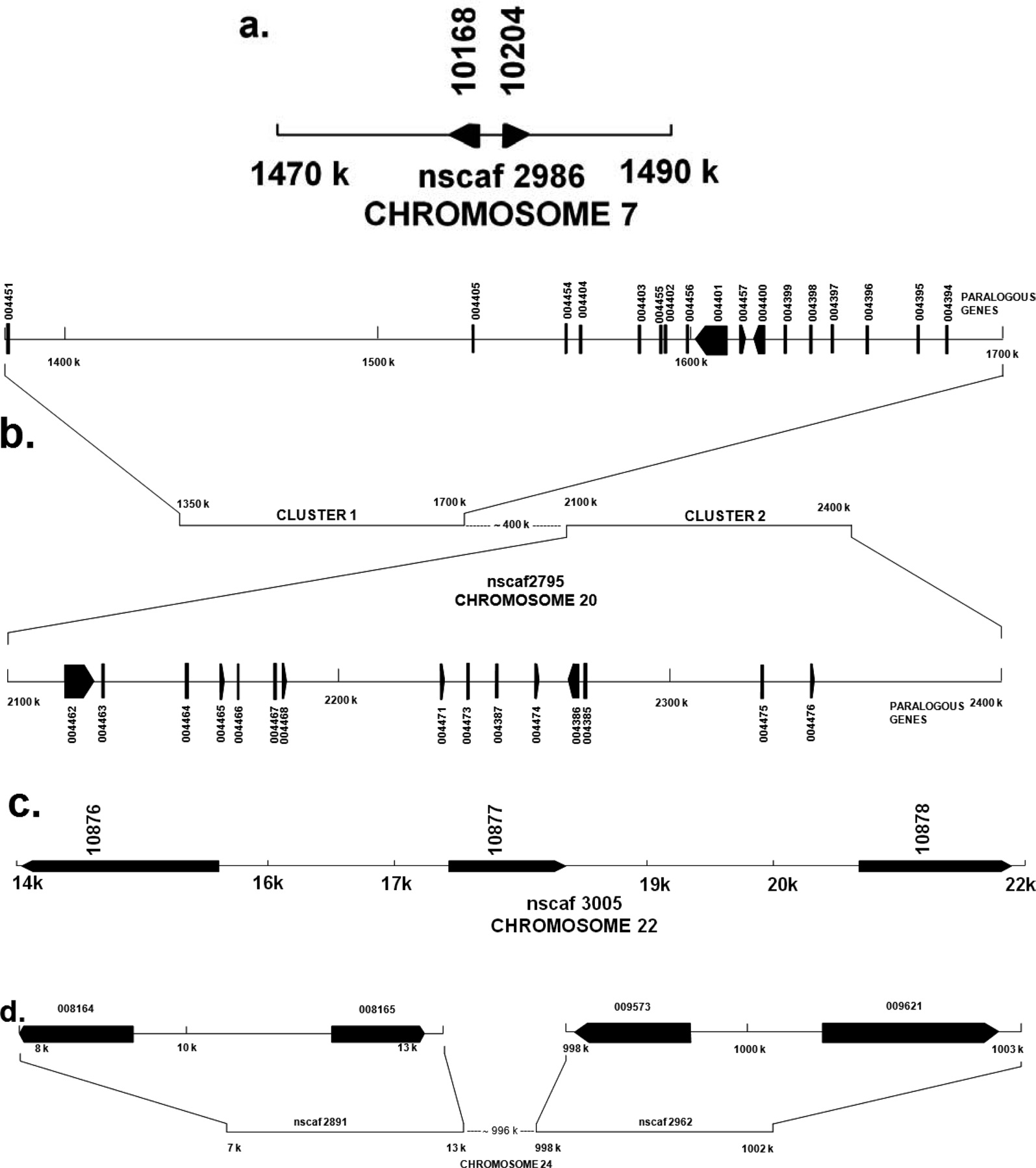
Genomic organization of B. mori PP-BP paralogous genes
The cDNA sequence of
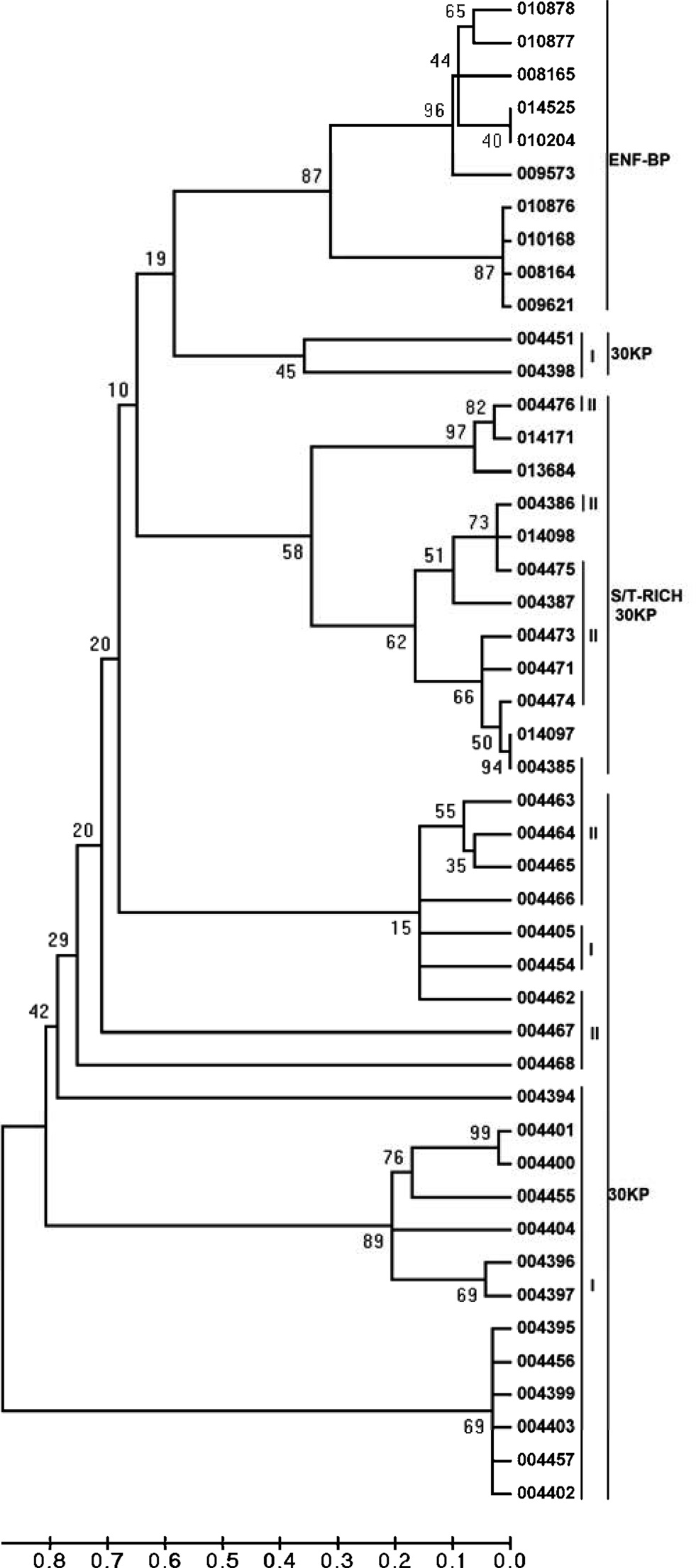
serine/threonine-rich 30KP. ClustalW analyses were performed with the multiple sequence alignment using ClustalW through MEGA 4 which revealed that these paralogous sequences formed three major clusters. Of the 46 sequences identified, 10 fall under the category of ENF-BP cluster, 12 fall under S/T-rich 30KP cluster and 24 of them fall under typical 30KPs clusters (Fig. 5).
Since the finding of the first member of the ENF peptide family, eleven lepidopteran species including
In the present study, the gene expression levels of PP and PP-BP were compared in diapause and nondiapause eggs after oviposition. The expression of PP was higher at 18 h similarly the expression of both PP and PP-BP genes was higher at 12 and 18 h along with few other genes in microarray experiment. These genes were taken up for further study, which were validated through q-PCR.
Most of the reports emphasize the role of ENF peptides in cellular immunity and cell proliferation. An
The structural and phylogenetic analyses of paralogous sequences revealed that these sequences can be categorized into three categories ENF-BP, typical 30k proteins and S/T rich 30K proteins. The 30k proteins are involved in the diapause mechanism through regulating the action of paralytic peptide.
In a study by Zhang
>
Multigene organization of B. mori PP-BP gene
Genes that have originated by gene duplication retained a certain degree of similarity forming multigene families. The multigene family members are often arranged in a compact cluster due to chromosomal rearrangements subsequent to gene duplications. The members of a multigene family can be functional or nonfunctional which are known as pseudo genes. Multigene families containing paralogous gene sequence that evolve in different ways, and assumption of the multigene family evolution is essential for estimation of its phylogeny. It is assumed that members of a multigene family may evolve at a faster rate and such members are designated as fast evolving genes. This phenomenon takes place when one gene member of a multigene family takes on a novel function, which is essential for survival and thus encounters significantly different selective pressure from other multigene family members. Another usual assumption of molecular tree of multigene family is that, each branch of the tree evolves independently from other branches. These families often show coincidental evolution, either indirectly through biased mutational and selective force or directly by mechanism such as gene conversion (Roach
A total of forty six
Similar phenomenon was observed for cecropin multigene family in B. mori. Cecropin B locus on genomic DNA consists of 6 paralogous gene groups as well as Cecropin D and E (Ponnuvel
In the current study it is observed that paralytic peptide is specifically up-regulated at 18h time point suggesting its possible role in induction of diapause along with other factors that contribute to the same. Excessive amount of PPs cause serious damage to insect self organization. Paralytic peptide binding proteins are also up-regulated during the diapause period, which suggests that PP-BP act as active regulators of the paralytic peptide (Matsumoto
Paralytic peptide and paralytic peptide binding protein interaction has to be further exploited to find out whether it is an alternative mechanism of diapause induction and also its possibility to design novel growth regulators in other insects.