Glycosylation is one of the most common post-translational modifications in biology.1 Effective and reliable analytical methods for glycans are important because the products of glycosylation are often present in clinical biomarkers.2 Recent developments in mass spectrometry (MS) have provided rapid and sensitive methods for the detection of a variety of glycans.3 Among the different matrices used in matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS, 2,5-dihydroxybenzoic acid (2,5-DHB) is one of the most common and is frequently used for analyses of synthetic polymers,4 peptides/proteins,5 and oligosaccharides.6 Recently, 2,6-DHB was introduced as an alternative matrix for the analysis of oligonucleotides, polysaccharides,7 and synthetic polymers.4 The combination of two matrix materials has been effective in several MALDI-TOF MS analyses. For example, a binary matrix of 2,5- DHB and α-cyano-4-hydroxycinnamic acid (CHCA) was effectively used to detect peptides and glycoproteins.8 Recently, binary matrices of 2,5-DHB/CHCA and 2,5-DHB/sinapinic acid were reported in successful analyses of intact glycoproteins and glycans.9
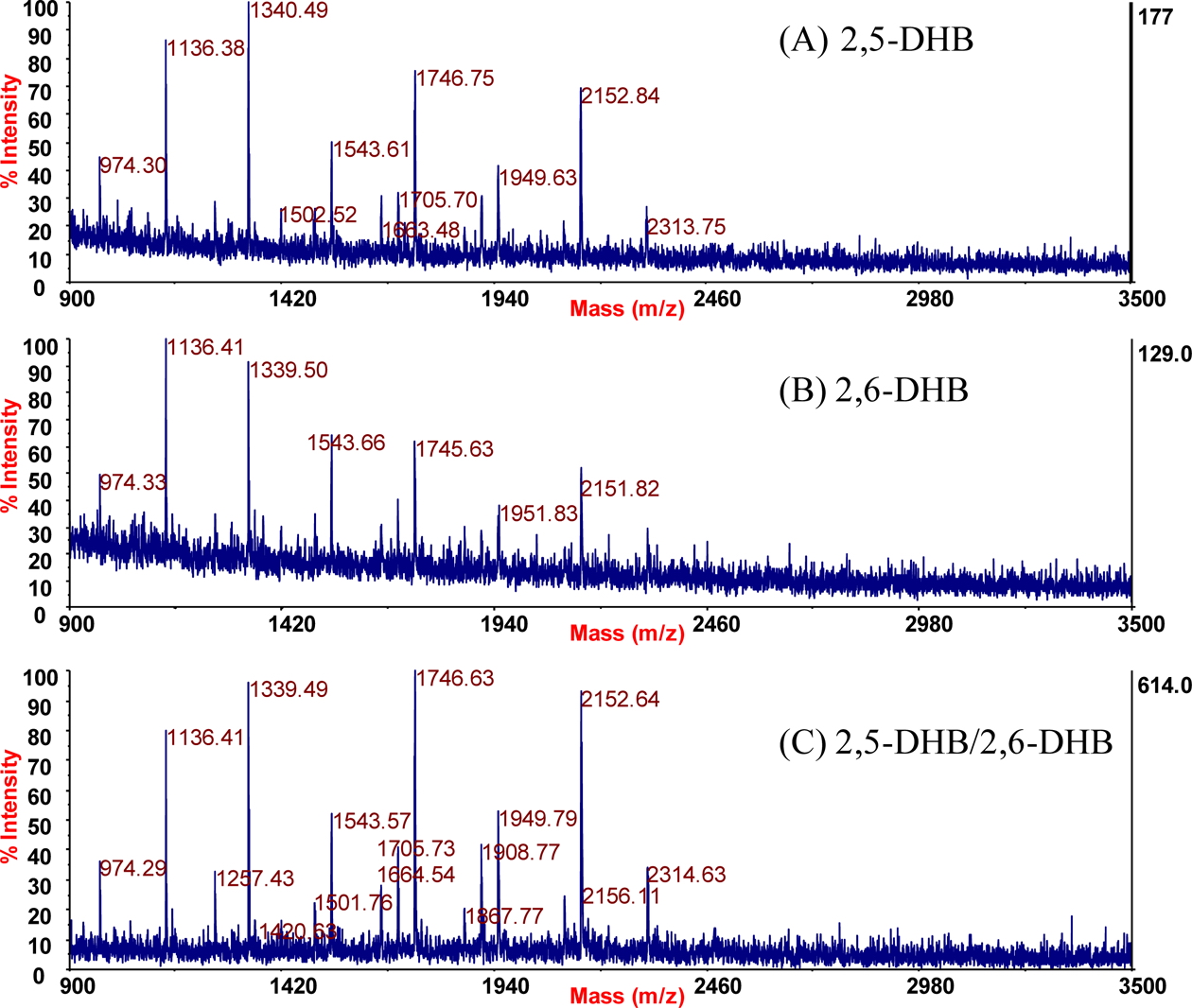
The current study investigated the effectiveness of a new binary matrix composed of 2,5-DHB and 2,6-DHB in analyses of glycans released from ovalbumin by enzymatic cleavage with PNGase F. The mass spectrum acquired with the binary matrix was compared with those obtained using the single matrix materials (2,5-DHB or 2,6-DHB).
All chemicals were obtained from Sigma?Aldrich (St. Louis, MO, USA) unless otherwise specified. To perform deglycosylation, 2mg of ovalbumin (catalog number: A5503) was mixed with 1 unit of PNGase F (catalog number: P7367) in 200 μL of 50mM ammonium bicarbonate buffer solution for 2 h at 37℃ with gentle mixing. To prepare the 2,5-DHB/2,6-DHB binary matrix solution, a mixture of 2,5-DHB (5 mg) and 2,6-DHB (5 mg) was dissolved in 1 mL of 50% acetonitrile (ACN) in H2O. For comparison, individual DHB matrices (2,5-DHB and 2,6-DHB) were also prepared to a concentration of 10 mg/ mL in 50% ACN. To prepare the sample targets for MALDIMS analysis, 0.6 μL of the matrix solution was loaded and allowed to dry on a MALDI plate and covered with 1 μL of the mixed sample/matrix solution (1:1, v/v). MALDI spectra were obtained on a Voyager MALDI-TOF mass spectrometer (Applied Biosystems) in reflection positive ion mode.
>
Improvements realized with the 2,5-DHB/2,6-DHB binary matrix
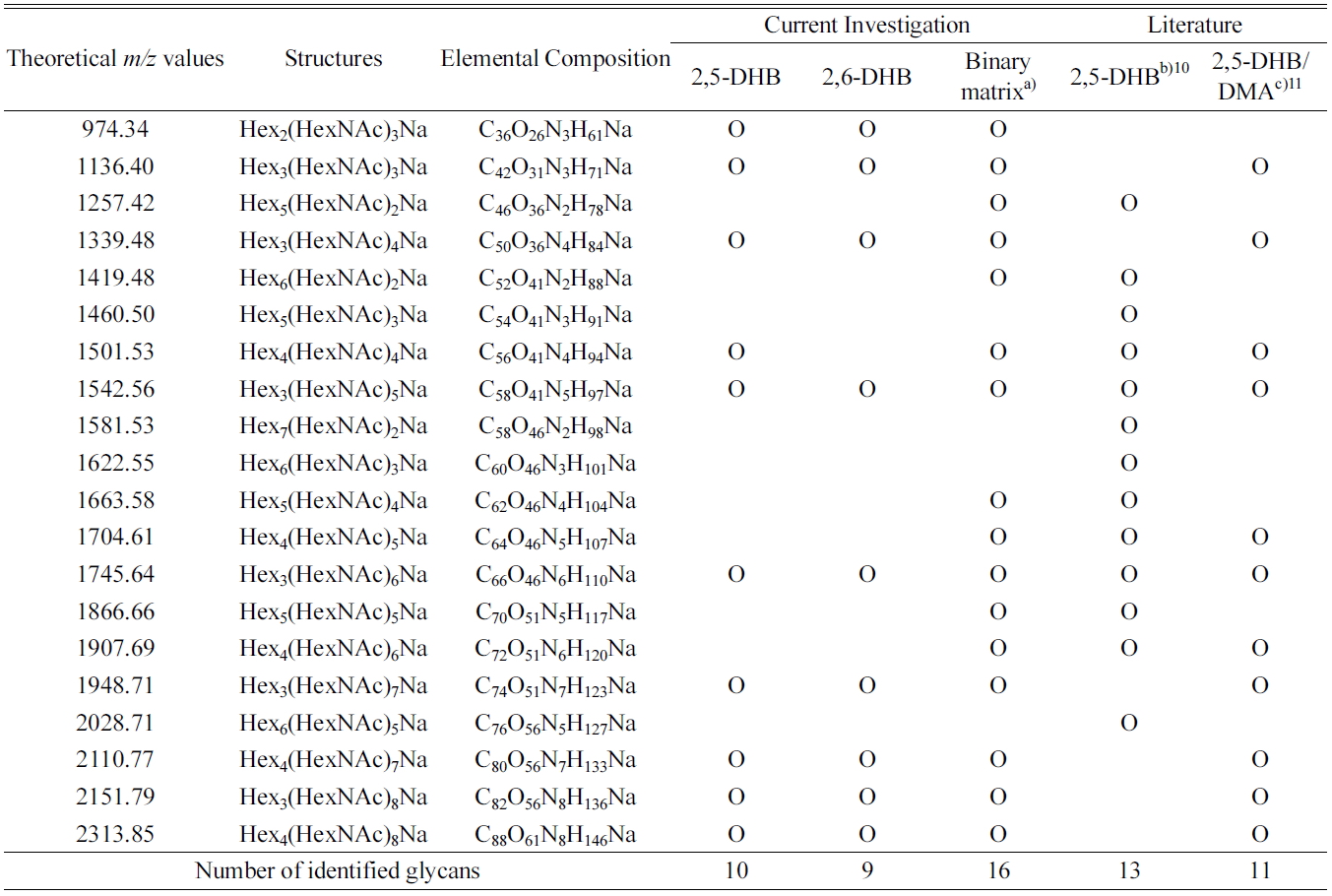
Figure 1 shows MALDI mass spectra of the glycans released from ovalbumin using PNGase F. Note that the highest number of glycans were detected using the binary matrix of 2,5-DHB/2,6- DHB. Table 1 summarizes the glycans identified in the current investigation along with the published results of other researchers. A total of 16 glycans were identified using the 2,5-DHB/2,6-DHB binary matrix, while only 10 and 9 glycans were identified when 2,5-DHB and 2,6-DHB were used independently, respectively. Harvey
[Table 1.] Summary of the identified glycans obtained from the deglycosylation of ovalbumin
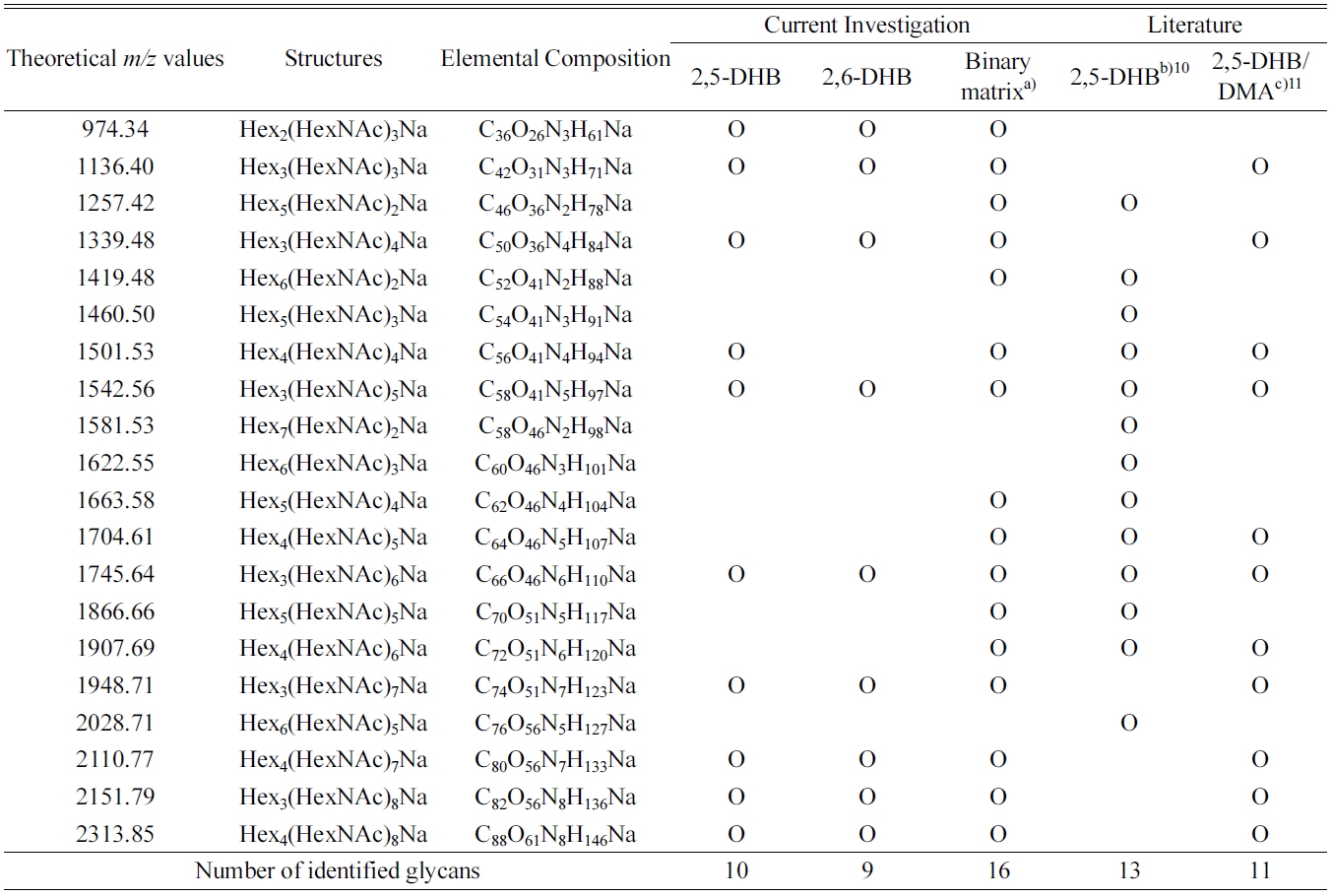
Summary of the identified glycans obtained from the deglycosylation of ovalbumin
(DMA) binary matrix to improve the MALDI detection efficiency of glycans. In that study, 11 glycans released from ovalbumin by PNGase F were identified, while no glycans were identified when using a 2,5-DHB matrix.11 Overall, the number of glycans observed using the 2,5-DHB/2-6-DHB binary matrix was higher than those detected using any other published matrix systems, including 2,5- DHB, 2,6-DHB, and 2,5-DHB/DMA.
The addition of phosphoric acid has been reported to enhance the signal intensities of phosphopeptides12 and nonphosphopeptides.13 Therefore, the addition of phosphoric acid was also investigated with regard to glycan analyses. No improvements were observed in any of the matrix systems used in the current study (2,5-DHB, 2,6-DHB, and 2,5-DHB/ 2,6-DHB). This is thought to be due to the fact that the current glycan detection requires sodium ion adducts in order to be detected by MALDI-MS. The effect of adding trifluoroacetic acid (TFA) to the matrix solutions was also investigated; no improvement was observed with all three matrix systems. Because the sodium adducts are the detected entity in positive ion mode MALDI-MS analysis of glycans, sodium cations, as NaTFA or NaCl, are often added to improve detection.6 However, the addition of NaTFA or NaCl to the sample solutions herein did not improve detection.
>
Reasons for enhanced signals
Uniform sample distribution within the MALDI sample spot, due to homogeneous crystallization,9 is believed to be the primary factor in realizing the observed improvement in glycan detection efficiency. In addition, synergistic effects of 2,5-DHB and 2,6-DHB, both of which have been reported as matrix materials in glycan analyses, are believed to enhance detection efficiency.
Various matrix materials (2,5-DHB, 2,6-DHB, and 2,5-DHB/ 2,6-DHB) were evaluated for their effectiveness in MALDI-TOF MS analyses of glycans. A 2,5-DHB/2,6-DHB binary matrix pr- ovided better performance than single matrices composed of 2,5- DHB or 2,6-DHB.