The damselfishes (Perciformes: Pomacentridae), which are widely distributed in tropical and warm temperate seas (Allen, 1991), comprise approximately 348 species in 28 genera (Nelson, 2006). The genus
Two specimens of
Total genomic DNAs were extracted from the muscle tissues of
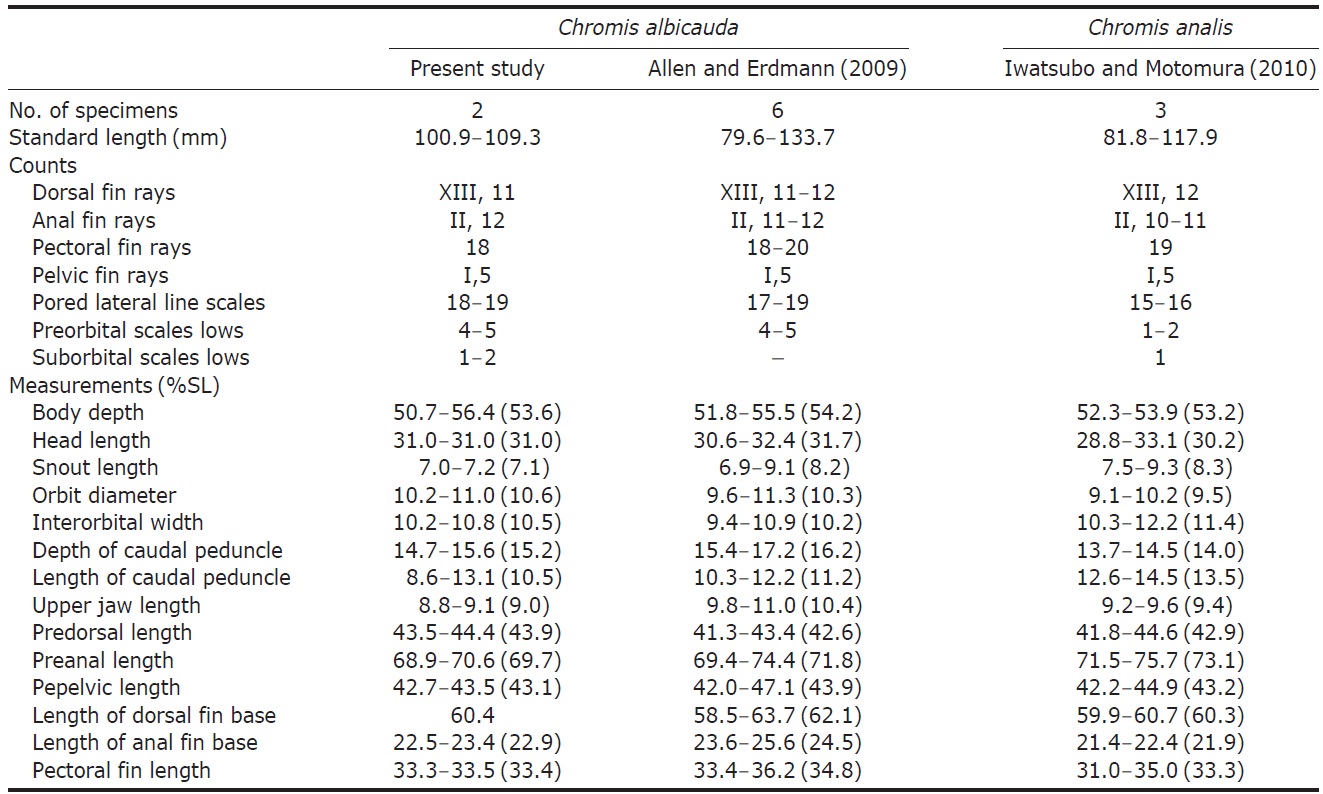
[Table 1.] Comparison of counts and measurements between Chromis albicauda and C. analis
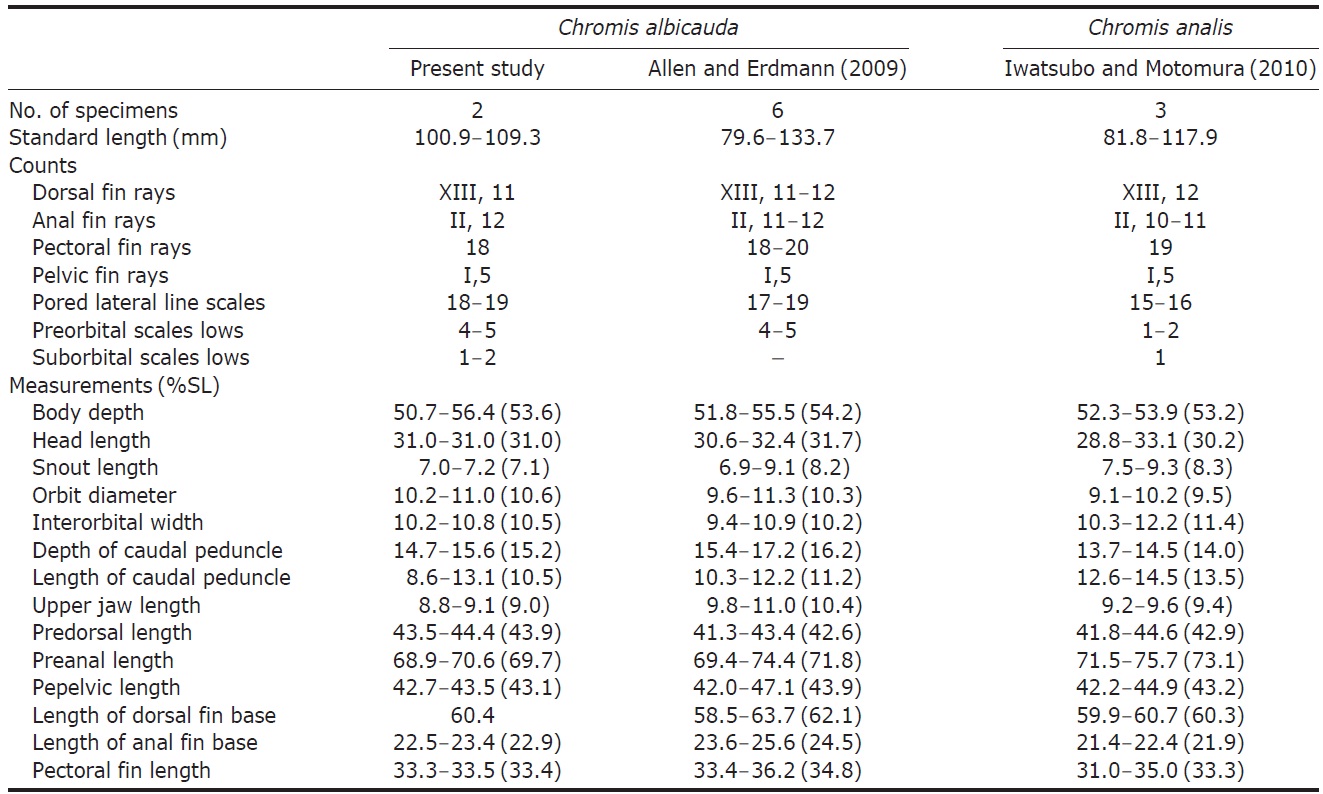
Comparison of counts and measurements between Chromis albicauda and C. analis
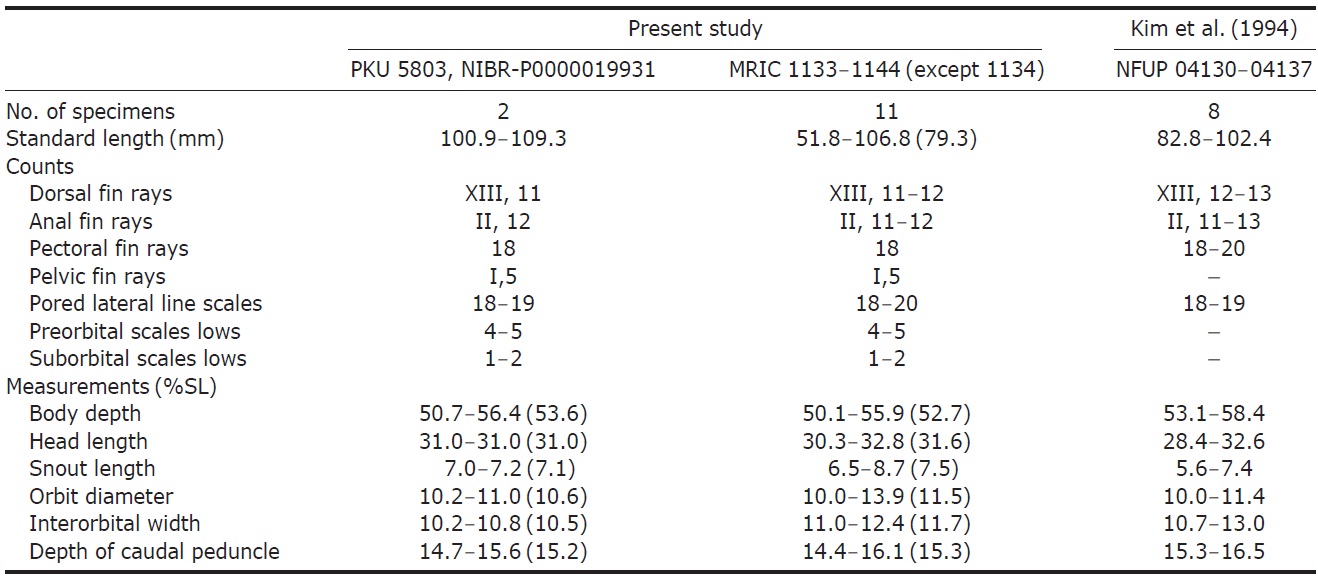
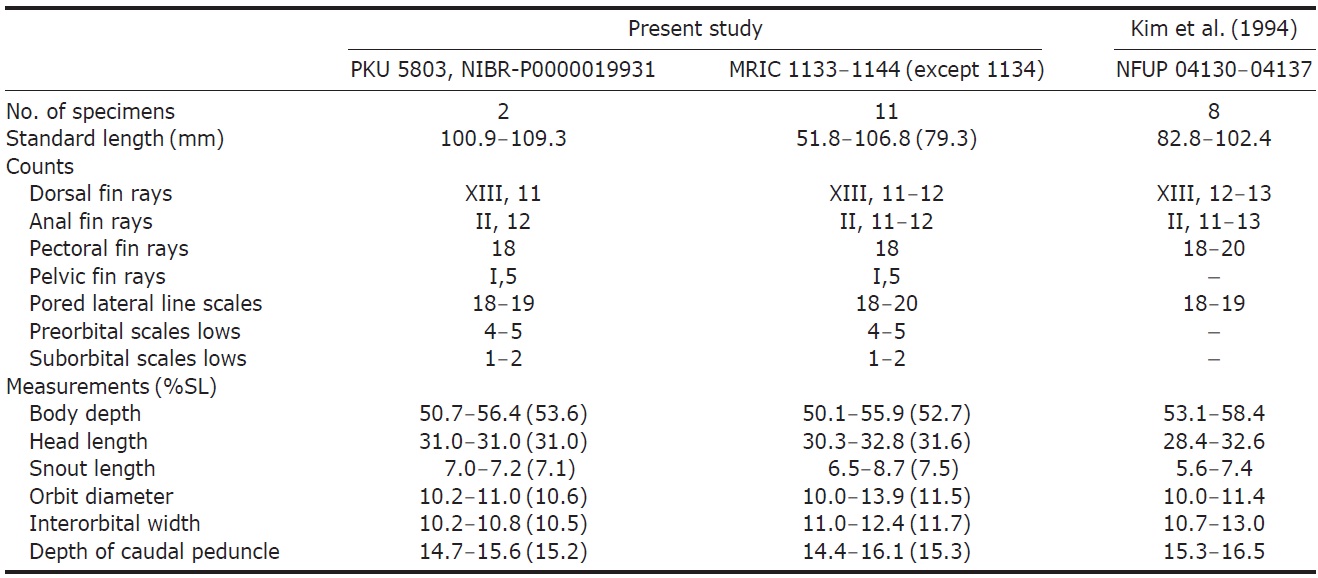
Comparison of counts and measurements of Chromis albicauda, among present study, compare with references
primers 16Sar-5′ (5′-CGC CTG TTT ATC AAA AAC AT- 3′) and 16Sbr-3′ (5′-CCG GTC TGA ACT CAG ATC ACG T-3′) (Ivanova et al., 2007). PCR was conducted on a thermal cycler (Bio-Rad MJ mini PTC-1148), with PCR solutions containing 10 μL of genomic DNA, 5 μL of 10× PCR buffer, 4 μL of 2.5 mM dNTP, 1 μL of each primer, 0.5 μL of FR taq polymerase (Biomedic, Korea), and 28.5 μL of distilled water. The PCR proceeded under the following the
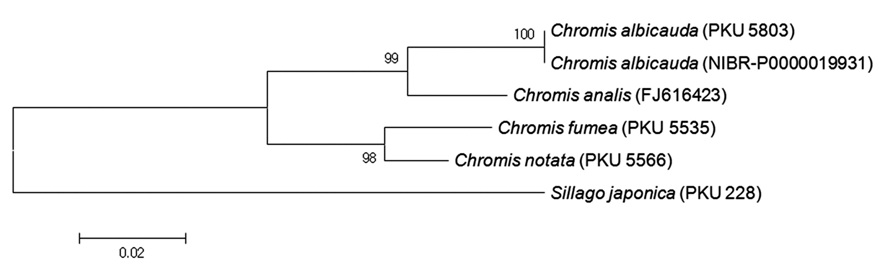
conditions: initial denaturation at 95℃ for 5 min, 35 cycles of denaturation at 95℃ for 1 min, annealing at 50℃ for 1 min, and extension at 72℃ for 1 min, and a final extension at 72℃ for 5 min. The amplification products of the target DNA were checked with agarose gel electrophoresis. The PCR products were purified with ExoSAP-IT (United States Biochemical Corp., USA). The DNA was sequenced using an ABI 3730XL sequencer and an ABI PRISMR BigDyeTM Terminator v3.0 Ready Reaction Cycle Sequencing Kit (Applied Biosystems, USA). Mitochondrial DNA 16S rRNA sequences were aligned and edited with Clustal W (Thompson et al., 1994) using BioEdit version 7.0.0 (Hall, 1999). The genetic distances were calculated according to Kimura two-parameter model (Kimura, 1980) using Mega 5 (Tamura et al., 2011). The neighbor-joining (NJ) tree was constructed using the Kimura two-parameter model (Kimura, 1980), where its confidence was assessed via 5,000 bootstrap replications. For molecular comparison, we further analyzed the mitochondrial DNA 16S rRNA sequences of the two
Order Perciformes
Family Pomacentridae
Genus Chromis
1*Chromis albicauda Allen and Erdmann, 2009 (Table 1, Fig. 1)
(new Korean name: Huin-ggo-ri-no-rang-ja-ri-dom)
Material examined. PKU 5803, 1 specimen, 109.6 mm in standard length (SL), Korea, Jeju-do, Seogwipo-si, Daejeongeup, Moseulpo, 9 Jun 2011, NIBR-P0000019931; 1 specimen, 100.9 mm SL, Jeju-do, Seogwipo-si, Daejeong-eup, Moseulpo, 23 Jun 2012.
Comparative materials examined. MRIC 1133-MRIC 1144 (except for MRIC 1134), 11 specimens, 51.8-106.8 mm SL, Korea, Jeju-do, Seogwipo-si, Bomok-dong, 26 Apr 2003 (all specimens were identified as
Description. D. XIII, 11; A. II, 12; P1. 18; P2. I, 5; LLp. 18-19; Vert. 25; GR. 24. Counts and measurements are shown in Tables 1 and 2.
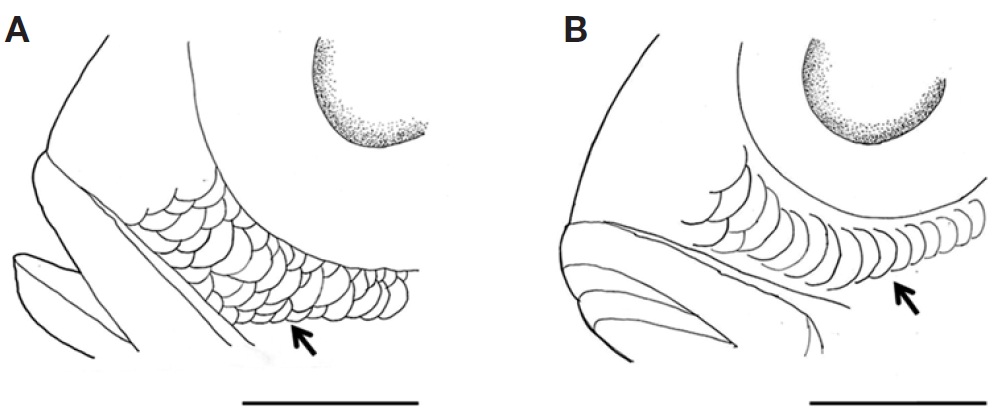
Body compressed; moderately deep, steeply inclined from frontal region to origin of dorsal fin. Mouth small and terminal. Snout shorter than orbit diameter. Posterior edge of maxilla reaching middle of orbit axis. Two pairs of nostrils; posterior nostrils larger than anterior nostrils. Conical teeth on both jaws. Well-developed sensory pores around eyes. Margin of preopercle extending to ventral region of jaw. Small scales on interorbital space; 4-5 irregular rows of small scales on the preorbital; 1-2 rows of relatively large scales on the suborbital (Figs. 1, 2). Operculum covered with large scales. Origin of dorsal fin vertically above upper axis of pectoral fin; dorsal spinous portion and soft portion smoothly connected. Pectoral fin, extending posteriorly to below 6th or 7th dorsal spine. Lateral line scales tubular, abruptly interrupted below the 1st dorsal soft ray. Caudal fin forked; three upper and lower procurrent caudal rays. Body covered with ctenoid scales; small scales present on bases of dorsal, anal, and caudal fins.
Coloration. When alive: overall yellowish; head dark-colored; small brown spot on origin of pectoral fin; caudal fin white (Fig. 1C). After fixation in formalin: overall yellowish tan; blackish blotch around anal opening (Fig. 1B); body dark brown dorsally and light yellowish ventrally; pectoral and pelvic fins yellowish tan, except dark brown at base of dorsal fins (Fig. 1A).
Distribution. Distributed in Indonesia (Allen and Erdmann, 2009), southern Japan (Iwatsubo and Motomura, 2010), and on the southern coast of Jeju Island, Korea (present study).
Remarks.
on the preorbital (see Tables 1, 2), and caudal fin color. According to the results of molecular analyses of 519 base pairs of mtDNA 16S rRNA sequences,
Korean name: 1*흰꼬리노랑자리돔