Accurate taxonomic information forms the basis of the protection and management policies for species conservation (Valdecasas and Camacho, 2003; Mace, 2004; Allendorf and Luikart, 2007). When a threatened species or subspecies is incorrectly classified and not legally protected, it can become extinct. Conversely, when common species are wrongly classified and overprotected, the budget and efforts that should be spent on endangered species may be wasted. The scops owl, designated as a natural monument (No. 324-7) and a protected species by the Korean Cultural Heritage Administration (CHA), is being threatened by many anthropogenic impacts collision, traffic accidents, habitat destruction, hunting and poisoning (Kim, 2006; CHA, 2010) finally, it is in the tendency of decreasing in population (The Korean Association for Bird Protection, 2012; CHA, 2010). Kim (2006) elucidated that collared scops owl is one of the most frequently rescued species accounted for 82.2% of avian records. Collision and traffic accidents were the main causes and especially traffic accident is the representative cause to threaten nocturnal raptor species. 93.7% of rescued natural monuments were raptors and 63.7% of the raptors were owl species including scops owl. Then, the need of counterplan to reduce traffic accidents of nocturnal raptors was suggested implying this scops owl might be placed in the serious circumstance without any conservation or management strategies. In spite of its importance, basic data for Korean collared scops owl is lack. Even though, this species is reported as both winter visitor and resident race (Deignan, 1950; Lee et al., 2000), this statement was not verified (The Ornithological Society of Korea, 2009). Besides, the use of the scientific name of the Korean collared scops owls has been inconsistent.
The scops owl occur in Korea were traditionally referred as Otus bakkamoena (see Cumming, 1933; Nam, 1950; Fennell, 1952; Vaurie, 1965; Dickinson, 2003; The Ornithological Society of Korea, 2009), though Austin (1948) described that the scops owls in Korea are common spring and autumn
transients which belong to Otus asio ussuriensis. On the other hand, Otus lempiji (Tomek, 1999; Lee et al., 2000) or Otus lettia (del Hoyo et al., 1999) were also used for the Korean scops owl. However, the distributions of these species are quite different (Fig. 1) (Weick, 2006). Otus lempiji is distributed in Southeast Asia, including the Malay Peninsula, the Southern Thai Peninsula, and Indonesia. Otus bakkamoena, known as the Indian or collared scops owl, is distributed in southeast Arabia and southeast Iran to southern Pakistan, part of Nepal, India, and Sri Lanka (Weick, 2006). The geographic range of Otus lettia (Hodgson, 1836) is from eastern Nepal to Sakhalin including Southeast Asian areas.
Based on its distribution within Korea, the existence of two populations of the scops owl has been also suggested in Korea: O. bakkamoena ussuriensis in the Korean Peninsula and O. bakkamoena semitorques on Jeju Island (Nam, 1950; Vaurie, 1965; The Ornithological Society of Korea, 2009). Therefore, some recent literatures also note these populations as two different species (O. lempiji ussuriensis and O. semitorques semitorques) (Clements, 2007) or even merge these groups into one single species O. lempiji (O. lempiji ussuriensis and O. l. semitorques) (Tomek, 1999) or O. semitorques (O. semitorques ussuriensis and O. s. semitorques) (Konig and Weick, 2008) which is distributed in southeast Siberia to Japan (del Hoyo et al., 1999; Weick, 2006; Fuchs et al., 2008). While there is record that Korean population has paler and grayer tones than Japanese population especially in its light markings (Deignan, 1950), when considering the geographic proximity and the possible existence of a local population belonging to Japanese population on Jeju island in Korea, the scops owl occurs in Korea may more closely related with O. semitorques rather than O. lempiji and O. bakkamoena. However, all the previous literatures on the Korean scops owl were based on its morphological features only. Therefore, in spite of the uncertainty and complexity on its status, the O. bakkamoena is the recommended name until confirmative studies on the phylogenetic status of the scops owl in Korea would be conducted (The Ornithological Society of Korea, 2009).
The phylogeny of Otus remains unclear (Heidrich et al., 1995; del Hoyo et al., 1999; Ryu and Park, 2003; Proudfoot et al., 2007; Fuchs et al., 2008). To resolve the species status of the Korean collared scops owls, a molecular phylogenetic study was conducted. We compared sequences of Korean collared scops owls, O. lempiji, with O. bakkamoena, O. lettia, and O. semitorques using two regions of mitochondrial DNA and confirmed the species status of Korean owls as well as relationships among the other species.
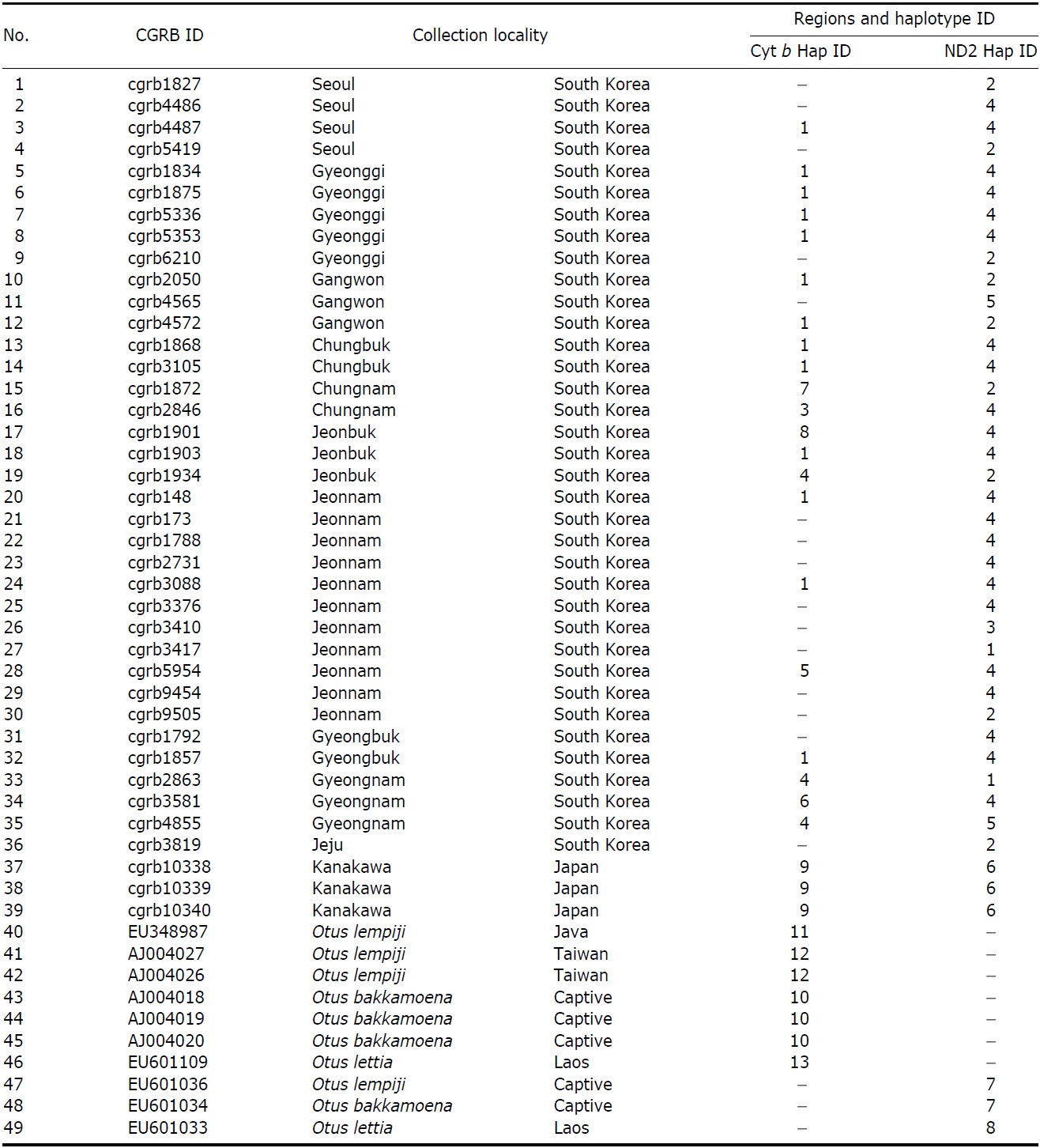
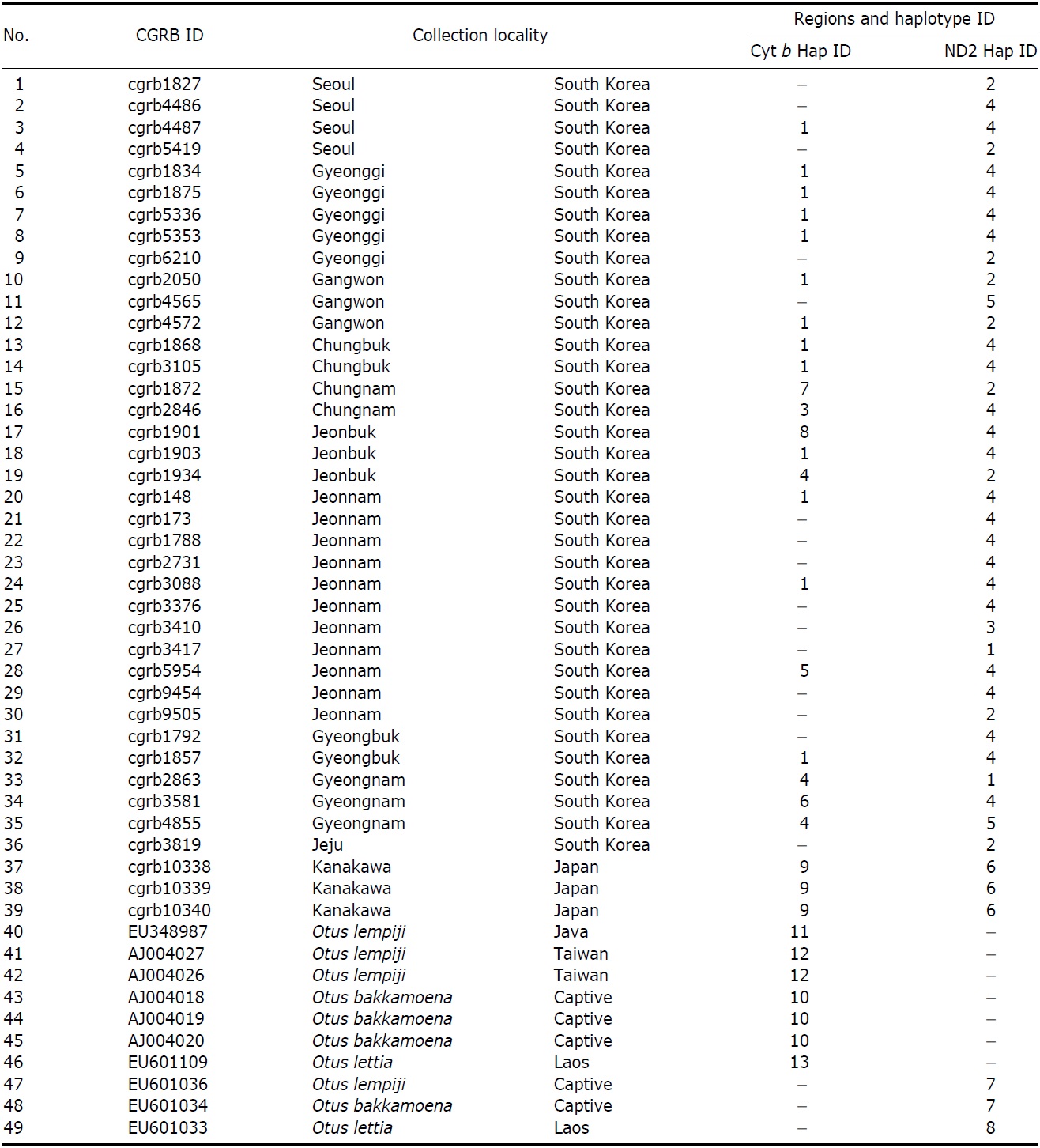
We obtained 36 tissue samples of Korean collared scops owls from the Conservation Genome Resource Bank for Korean Wildlife (CGRB; CGRB constructed a network of wildlife rescue centers and animal hospitals where injured or dead wild animals are brought throughout Korea). We obtained three DNA samples of O. semitorques (from bird strikes or traffic accidents) from Japan. The sequences of the other Otus species, O. lempiji (EU348987, AJ004026-7 for cytochrome b [cyt b]; EU601036 for NADH dehydrogenase subunit 2 [ND2]), O. bakkamoena (AJ004018-20 for cyt b; EU601034 for ND2) and O. lettia (EU601109 for cyt b; EU601303 for ND2) for phylogenetic analysis, were obtained from GenBank (Table 1).
DNA was extracted using DNeasy Tissue and Blood Kits (Qiagen, Valencia, CA, USA). Two mitochondrial regions were amplified using the following primers: 1) cyt b gene using L14841 (Kocher et al., 1989) and H16065 (Brito, 2005) and 2) ND2 gene using L5219 and H6316 (Meng et al., 2008). Each 30 μL PCR reaction contained ~50 ng of template DNA, 1.5 mM MgCl2, 2.5 mM dNTPs, and 1 U (0.2 μL) of i-star Taq polymerase (5 U/μL, iNtRON Biotechnology, Seongnam, Korea). Thermocycling conditions started with an initial denaturation of 94℃ for 5 min, followed by the touch-down method of 20 cycles of 45 s at 94℃ for denaturation, 45 s at 55℃ for annealing (decreasing 0.5℃ per cycle to 45℃), and 45 s at 72℃ for extension, and 25 more cycles of 45 s at 94℃
for denaturation, 45 s at 45℃ for annealing, and 45 s at 72℃ for extension, ending with a final extension of 10 min at 72℃. PCR products were purified using a Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA, USA) and directly sequenced in both directions using ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems Inc., Carlsbad, CA, USA), followed by detection on an ABI 3730 XL DNA sequencer.
The sequences were edited by Geneious Pro version 4.7 (Drummond et al., 2009) and aligned using Clustal X (Thompson et al., 1997). Phylogenetic trees were constructed using maximum-likelihood (ML) and maximum-parsimony (MP) methods by PAUP version 4.0 beta 10 (Swofford, 2001). The MP tree was constructed from 100 replicates using a heuristic search algorithm with tree-bisection-reconnection. In the ML analysis, HKY85+G model for cyt b and HKY85 model for ND2 were chosen as the best fit model by Modeltest 3.7 (Posada and Crandall, 1998) and heuristic search of 100 replicates was carried out. The neighbor-joining (NJ) method was also used to construct a phylogenetic tree with 1000 bootstrap replicates by MEGA version 4 (Tamura et al., 2007). The number of polymorphic sites (S), haplotype diversity (h), and nucleotide diversity (π) were calculated using DnaSp 4.10 software (Rozas et al., 2003). Pairwise sequence distances were calculated based on the Kimura two-parameter method (Kimura, 1980).
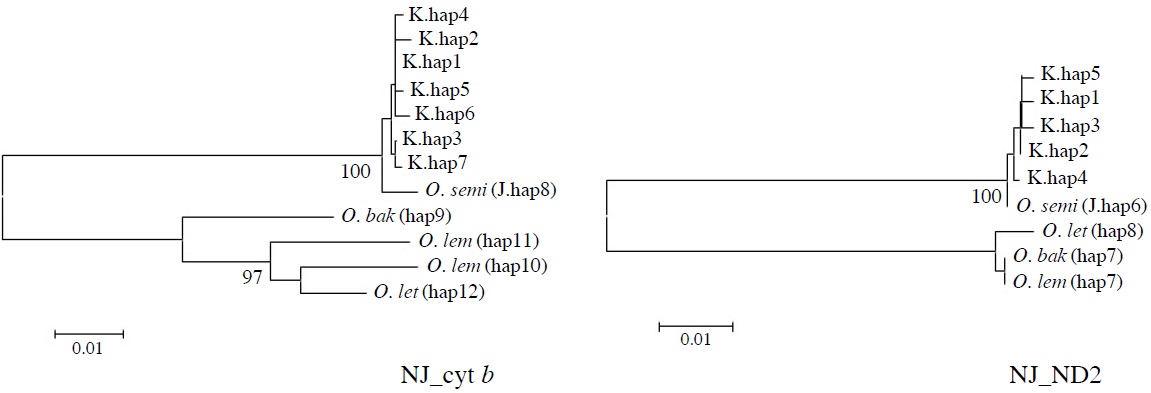
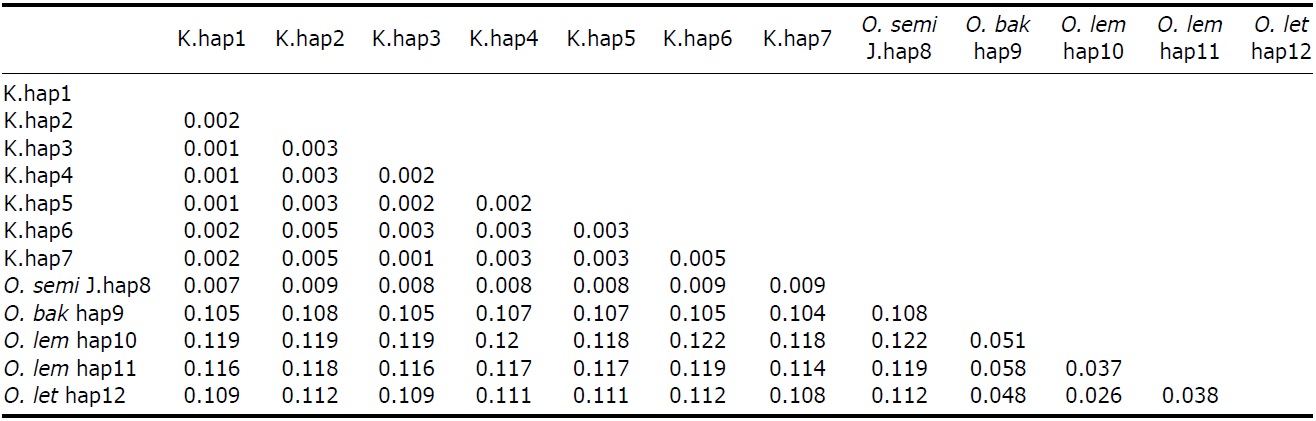
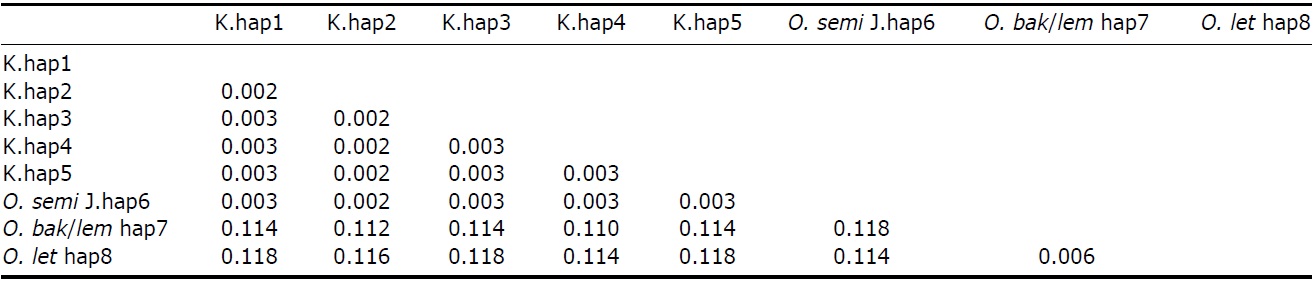
[Fig. 2.] Molecular phylogenetic trees of Korean collared scops owls, Otus lempiji (O. lem), O. bakkamoena (O. bak), O. semitorques (O. semi), and O. lettia (O. let). All maximum-likelihood (ML), maximum-parsimony (MP), and neighbor-joining (NJ) trees revealed same aspects. NJ tree of cytochrome b (cyt b, 891 bp) and NADH dehydrogenase subunit 2 (ND2, 627 bp) using haplotypes was showed (not shown ML and MP trees). K, Korean; J, Japanese.
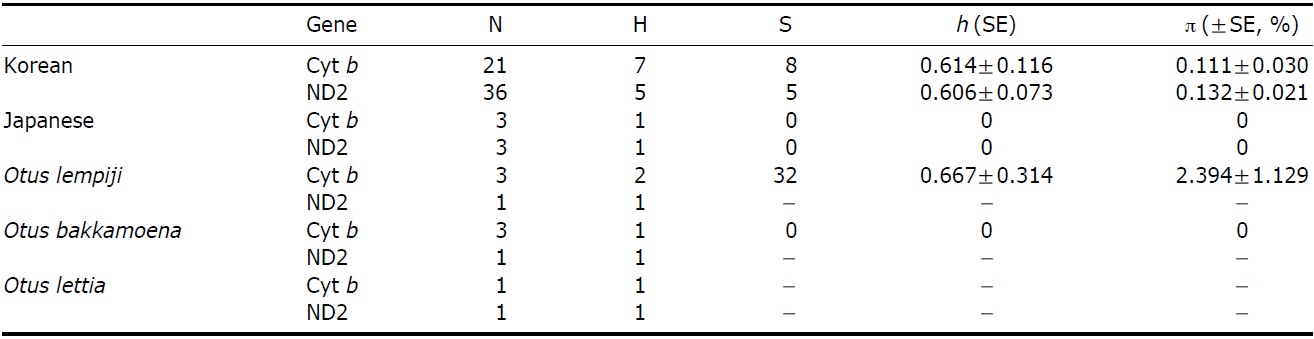
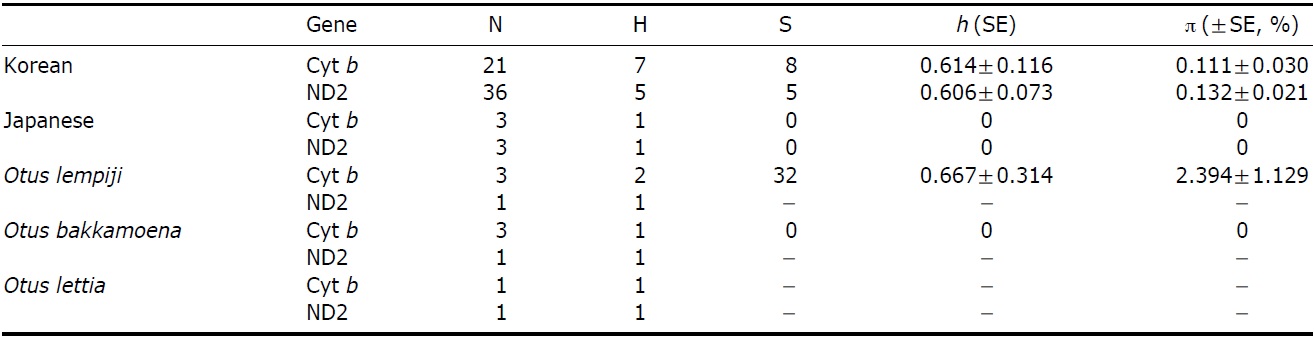
The partial sequences of cyt b (891 bp, JN11958-81) and ND2 (627 bp, JN11982-12020) were successfully determined from 24 cyt b and 39 ND2 collared scops owl specimens. We compared the sequences with O. lempiji (EU348987, AJ004026-7 for cyt b; EU601036 for ND2), O. bakkamoena (AJ004018-20 for cyt b; EU601034 for ND2) and O. lettia (EU601109 for cyt b; EU601033 for ND2) from GenBank. There were no indels present in either gene. In the cyt b gene, there were eight polymorphic sites, resulting in seven haplotypes. ND2 sequences showed five polymorphic sites and five haplotypes. The overall haplotype diversity and nucleotide diversity of Korean specimens were 0.614±0.116 and 0.111±0.030%, respectively, in cyt b and 0.606±0.073 and 0.132±0.021%, respectively, in ND2 (Table 2).
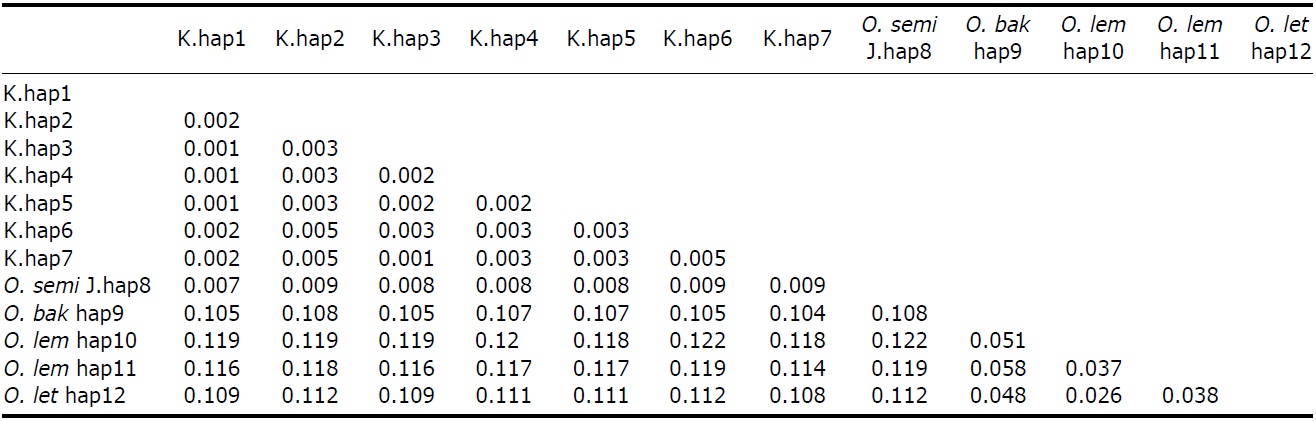
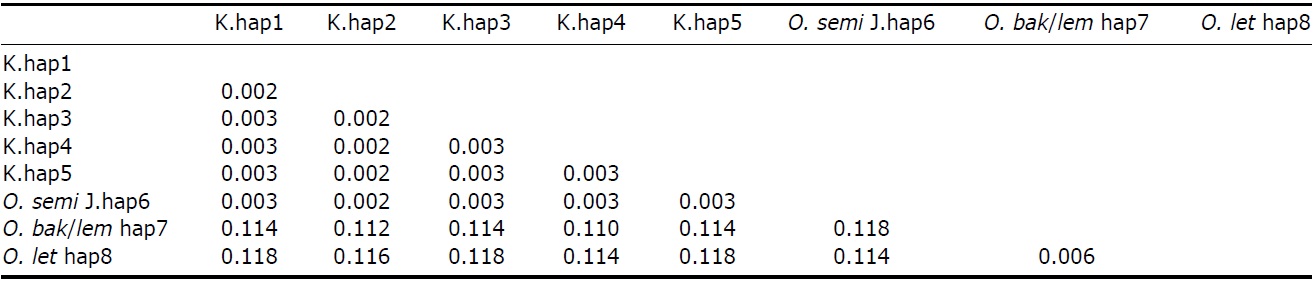
All ML, MP, and NJ trees of cyt b and ND2 sets were very similar to each other, with only minor differences. Through phylogenetic analysis, we confirmed that trees in both regions consisted of two clades with 100% bootstrap values: Clade 1, Korean species and O. semitorques; Clade 2, O. lempiji, O. bakkamoena and O. lettia (Fig. 2 representative NJ tree; ML/MP trees not shown). All the Korean collared scops owl haplotypes were grouped together with very low genetic differences (cyt b, d=0.001-0.005; ND2, d=0.002-0.003) and were allied with O. semitorques (cyt b, mean d=0.004; ND2, mean d=0.003) (Tables 3, 4). When compared with O. lempiji, the genetic distance between the Korean samples and O. lempiji was extremely high (cyt b, d=0.118; ND2, d=0.113) (Tables 3, 4) and could be considered a species level difference. We also found a high genetic difference between the Korean species and O. bakkamoena (cyt b, d=0.106; ND2, d=0.113)/O. lettia (cyt b, d=0.110; ND2, d=0.117), respectively (Tables 3, 4). However, moderate (cyt b, 0.043) or very low (ND2, 0.003) genetic differences were observed among O. lempiji, O. bakkamoena and O. lettia, even, no genetic difference of ND2 gene between O. lempiji and O. bakkamoena was shown. No regional separation was observed among the Korean samples.
The collared scops owl, a natural monument and protected species of Korea, is the subject of questions concerning its formal scientific name and phylogenetic status. The taxonomic status of this species with other related Otus species is not clear. Several studies have been performed to resolve this issue (Heidrich et al., 1995; Wink and Heidrich, 2000;
Ryu and Park, 2003; Wink et al., 2004; Fuchs et al., 2008), but these efforts highlighted other Otus species or related species without including the Korean collared scops owl. As a consequence, O. lempiji, O. bakkamoena, O. lettia, and O. semitorques are still of ambiguous phylogenetic status (Konig et al., 1999; Weick, 2006; Fuchs et al., 2008; Lok et al., 2009). Their population has been decreasing gradually owing to many anthropogenic factors especially collision and traffic accidents (Kim, 2006; CHA, 2010). Moreover, if these threatening factors are not removed, the Korean species may become endangered. At this point, unclear classification could be an obstacle to conservation strategies. The aim of the present study was to clarify the scientific name, to clear the confusion surrounding the taxonomic status of the Korean collared scops owl, and to provide basic information for its conservation. We also sought to verify relationships among the three Otus species using mitochondrial cyt b, which has been used for owl systematic classification (Heidrich et al., 1995; Griffiths, 1997; Mindell et al., 1997; Ryu and Park, 2003; Wink et al., 2004), and additional ND2 sequences.
All the ML, MP, and NJ trees of both regions (cyt b and ND2) confirmed that the Korean species is clearly different from O. lempiji and O. bakkamoena, which are the scientific names used officially for the collared scops owl in Korea. Instead of clustering with O. lempiji, O. bakkamoena, or O. lettia, they aligned with the morphologically similar species, O. semitorques (Fig. 2). This implies that the Korean species is the same as O. semitorques. Further genetic and ecological data are necessary to verify this association. A slight genetic difference between the Korean collared scops owl and O. semitorques was detected in cyt b gene (Table 3), which may be considered as a regional variation (Deignan, 1950; Weick, 2006).
In the cyt b gene, O. lempiji (hap11) was allied with O. lettia
more closely than another O. lempiji (hap12), moreover, in the ND2 region, O. lempiji and bakkamoena had the same sequence conformation (Tables 3, 4, Fig. 2), which may be due to the fact that the length of the sequences was not long enough to discriminate between the two species. Alternatively, sampling error can be another explanation. O. lempiji is often considered a race of O. bakkamoena or O. lettia (Weick, 2006) and members of the genus Otus are similar in plumage. Species discrimination of some taxa relies only on vocalization (Weick, 2006; Fuchs et al., 2008); therefore, individuals described as O. lempiji and O. lettia (cyt b)/O. bakkamoena (ND2) may be of the same species.
We had similar results with the MHC II gene (534 bp, nuclear DNA, unpublished to date), which grouped Japanese specimens together with Korean collared scops owls with very low genetic differentiation (d=0.004). No regional difference between Korean and Japanese specimens was detected. According to previous Otus species studies, mitochondrial genetic difference value among different species was 0.031- 0.186 (Heidrich et al., 1995; Ryu and Park, 2003). Thus, we confirmed that the genetic distance between the Korean samples and O. lempiji/bakkamoena/lettia falls within the general interspecific differentiation range (Tables 3, 4).
Justification for changing the scientific name of the Korean collared scops owl to O. semitorques is supported by the following evidence: First, Deignan (1950) mentioned that the Korean species defined as O. b. ussuriensis seems to be inseparable from the Japanese specimens based on plumage color and wing length, and suspected that semitorques would prove to be the resident race of Southern Korea; second, there was a phylogenetic study of the family Strigidae which referred to the Korean species as semitorques (Ryu and Park, 2003) and also indicated unclear divergence among Otus species; third, in the Owls (Weick, 2006), we can definitely confirm the Korean scops owl is systematically classified as a subspecies of semitorques. According to Weick (2006), O. lempiji, O. bakkamoena, O. lettia, and O. semitorques are distributed in different areas. Moreover, lempiji is frequently considered as O. bakkamoena or O. lettia, although they have different vocalizations. In the case of O. semitorques, it is often regarded as conspecific with the morphologically variable O. bakkamoena; however, they differ in vocalizations and the color of irises (O. semitorques has red irises). Finally, Konig and Weick (2008) described Korean species as O. semitorques in recent years.
Since owls are nocturnal, their morphological characteristics are hard to observe so classification can be overlooked (Wink et al., 2004). Even though half of the Otus taxa are recognized as different species according to morphological traits, other species are still a matter of debate (Heidrich et al., 1995). Thus far, distinctive vocalization is used as a major key for discriminating between Otus species (Hekstra, 1982; Konig, 1991, 1994). If genetic information is added to morphological and vocal characteristics, phylogenetic relationships of owls will be better defined. Research of Otus atricapillus complex using mitochondrial DNA is a remarkable example to define four independent species which are morphologically very similar, have been considered as a single species (Heidrich et al., 1995). Even though there is abundant data to support the reclassification of the Korean collared scops owl as O. semitorques, there have not been any studies verifying this information to confirm that the incorrect scientific name has been used. We performed molecular analysis with mitochondrial DNA and acquired persuasive results. Based on our study, we suggest that the formally used scientific name of the Korean collared scops owl should be changed to Otus semitorques and this finding will be used as the basis for developing effective conservation or management strategies.