The genus
Blooms of cyanobacteria are not only a significant problem in terms of water quality, but also pose a severe risk to human and animal health. They may contain potent hepato- and neurotoxic, as well as cytotoxic agents produced by strains of several cyanobacterial genera. It has been documented that dogs, birds and young cattle have all been killed by hepatotoxins over the years and forty cows suffered from fatal neurotoxin poisoning at Lake Vesijarvi, Finland in 1928 (Sivonen 2009).
Paddy field ecosystems provide an environment favorable for the growth of heterocytous cyanobacteria due to the moderate light, water, high temperature, and nutrient availability. The paddy fields in question are submerged for 60-90 days during the growing season (Roger and Kulasooriya 1980).
Most researchers in the fields of cyanobacteria toxicology are of the opinion that the toxic compounds produced by toxic aquatic cyanobacteria may leak into the submerged paddy fields when the algae lyse.
As there have been no reports of toxicity from drinking water contaminated with aquatic cyanobacteria in the submerged paddy fields, we decided to conduct a preliminary study of toxic chemicals in order to identify the possible causative compounds of this toxicity in one strain of heterocytous cyanobacteria which is frequently found in paddy fields of Iran.
Discovery of the presence of three diverse groups of potential bioactive compounds with strong anti-fungal and toxigenic properties in the dominant
In 2010, soil samples were collected from five paddy fields in Golestan province, Iran. Five grams of soil from each sample were weighed and aseptically transferred to sterile petri dishes with adequate quantities of liquid media BG110 (Rippka et al. 1979) without NaNO3. The pH was adjusted to 7.1 after sterilization. The petri dishes were incubated in a culture chamber at 28℃, and were provided with continuous artificial illumination of approximately 1,500-2,000 lux for two weeks (Kaushik 1987). Three small fragments of growing colonies were then placed equidistantly onto an agar surface for purification. Morphological measurements were made by bright-field microscopy and by phase-contrast illumination of 10-day-old cultures using a Leica DM750 P microscope (Leica, Wetzlar, Germany). The following parameters were selected to describe the morphology of heterocytous cyanobacteria: morphology of vegetative cells (including terminal cell), heterocytes, akinetes; presence or absence of terminal heterocytes; and the shape of the filament and its aggregation in colonies (Rajaniemi et al. 2005). The species were identified according to Desikachary (1959).
One strain of heterocytous cyanobacteria (ASN_M) which was the most frequently observed species in the paddy fields was selected for evaluation of its toxigenic potential, bioactivity and chemical analysis. ASN_M was grown in liquid media BG110 (Rippka et al. 1979). One milliliter of culture was transferred to a sterile 1.5 mL microtube and was stored at -20℃ prior to anatoxin analysis. The remaining cell mass was harvested by centrifugation at 7,000 ×g for 7 min, resulting in a cell mass of 336 mg (wet weight).
The harvested biomass was transferred to two separate sterile 1.5 mL microtubes, each containing 50 mg wet mass for DNA extraction, and the remaining 236 mg for chemical and bioactivity analyses.
The extract for the anatoxin analysis was prepared from 1 mL frozen culture. The microtube containing the culture was placed in a water bath at 23℃ to thaw. Subsequently, it was supplemented with 300 mg glass beads (disruptor beads, 0.5 mm; Scientific Industries, Bohemia, NY, USA) and 2 μL 50% (v/v) formic acid (Fluka/Sigma- Aldrich, Steinheim, Germany) (final concentration 0.1% [v/v] formic acid). The cells were broken mechanically three times for 15 s at a speed 6.5 m s-1 with a FastPrep instrument (Savant Instruments Inc., Holbrook, NY, USA). The homogenised mixture was centrifuged at 10,000 ×g for 5 min. Twenty microliters of the supernatant was diluted 1 : 10 with acetonitrile (Chromasolv for LC/MS; Fluka/ Sigma-Aldrich), equalling to a final volume 200 μL and the extract was then analysed for chemical analysis by liquid chromatography mass spectrometry (LC-MS) using an Agilent 1100 Series LC/MSD Trap XCT Plus System (Agilent Technologies, Palo Alto, CA, USA).
For the preparation of methanol extracts, 236 mg wet biomass was freeze-dried (Edwards lyophilisator) yielding a dry cell mass of 7.3 mg. The entire dry cell mass was added to a microtube along with 300 mg glass beads (disruptor beads, 0.5 mm; Scientific Industries) and 1 mL methanol (LC-MS grade; Fischer Scientific, Pittsburg, PA, USA) was homogenised three times for 15 s at a speed of 6.5 m s-1 with a FastPrep instrument (Savant Instruments Inc.) The mixture was then centrifuged as described above. The supernatant was stored at 4℃ until chemical and bioactivity analyses were performed. For the detection of microcystin, the supernatant was diluted 500 times with methanol (LC-MS grade; Fischer Scientific).
Genomic DNA of ASN_M was extracted utilising the E.Z.N.A. SP Plant DNA kit (Omega Bio-tek, Inc., Norcross, GA, USA). The microtube containing 100 mg wet cells was supplemented with 300 mg of two differently sized glass beads (acid-washed, 180 μm and 425-600 μm; Sigma- Aldrich) as well as lysis buffer and RNase solution, both provided by the kit. In order to ensure proper disruption of the cells, tubes were homogenised three times for 20 s at a speed of 6.5 m s-1 with a FastPrep instrument (Savant Instruments). The extraction procedure was continued according to the kit’s protocol, as supplied by the manufacturer. DNA was quantified with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, USA).
16S rRNA gene-based identification
Polymerase chain reaction (PCR) amplification. Two oligonucleotide primers comprised of one forward primer (359F, 5'-GGGGAATYTTCCGCAATGGG-3') and a reverse primer (781Ra, 5'-GACTACTGGGGTATCTAATCCCATT-3' ) (Nubel et al. 1997) were used for partial amplification of 16S rRNA gene.
One PCR reaction was comprised of 1 time buffer solution (DyNAzyme PCR buffer; Finnzymes, Espoo, Finland), 0.5 μm forward primer, 0.5 μm reverse primer and 0.5 U Taq polymerase (DyNAzyme II DNA polymerase; Finnzymes) as well as 1 μL template DNA and sterile water in a total volume of 20 μL. The template DNA concentration of the ASN_M strain in the reaction accounted for approximately 140 ng.
As a positive control for the amplification procedure, genomic DNA of the cyanobacterium
PCR products were checked by electrophoresis on 1% agarose gels (SeaPlaque GTG; Cambrex Corp., East Rutherford, NJ, USA) at 100 V, followed by 0.10 μg mL-1 ethidium bromide (EtBr; Bio-Rad) staining.
PCR products were visualised in the gel by UV light utilising the Molecular Imager Gel Doc XR system (Bio-Rad). A digital gel image was obtained utilising the imageanalysis software (software BioRad/Quantity One version 4.6.7).
The size of the products was estimated by comparison to marker DNA (λ/HinfIII + φx/HaeIII; Finnzymes). The products were purified using the Geneclean Turbo kit (Qbiogene/MP Biomedicals, Solon, OH, USA) and werequantified with a Nanadrop ND-1000 spectrophotometer(Thermo Scientific, Wilmington, DE, USA).
Sequencing and analysis. Sequencing of the partial 16S rRNA gene of ASN_M strain was subsequently carried out by the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Life Technologies, Foster City, CA,USA) with 10 μL reaction mixtures each containing 2 μL sequencing buffer, 1 μL big dye, 1 μm primer and 42 ng PCR product. Reactions for forward and reverse primers were prepared separately.
BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST) of the partial 16S rRNA gene of ASN_M strain were used to identify similar sequences deposited in the GenBank database of NCBI. The 16S rRNA gene sequences obtained in this study, as well as reference sequences retrieved from GeneBank were first aligned with CLUSTAL W with the default settings and were then manually edited in BioEdit version 7.0. The positions with gaps, as well as undetermined and ambiguous sequences were removed for subsequent phylogenetic analyses. Phylogenetic trees using the neighbour-joining method was constructed by the MEGA version 4.1 (Tamura et al. 2007) using the Kimura two-parameter model. The robustness of the tree was estimated by bootstrap percentages using 1,000 replications. The root of the tree was determined using the 16S rRNA of
>
PCR amplification of potential toxin genes
To determine if the ASN_M strain is a potential produc-
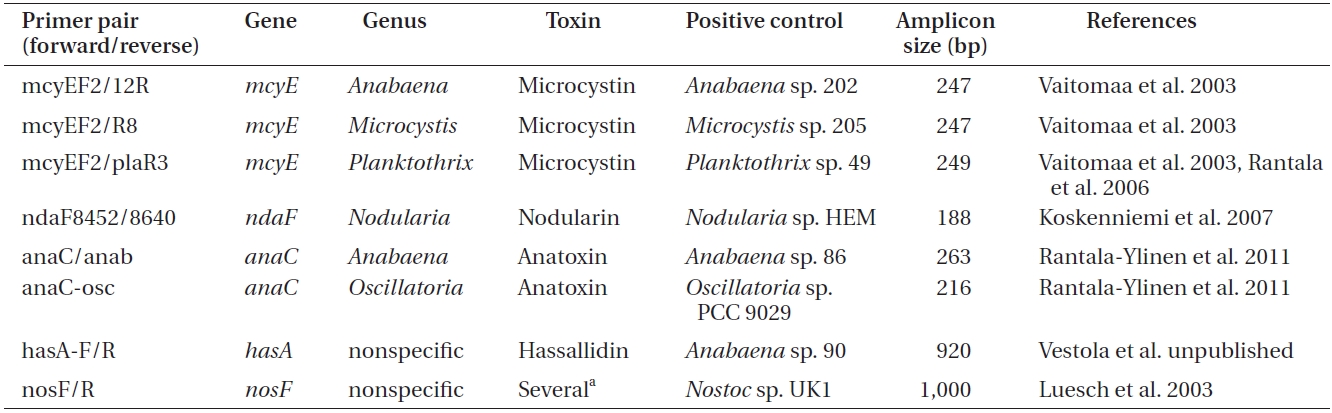
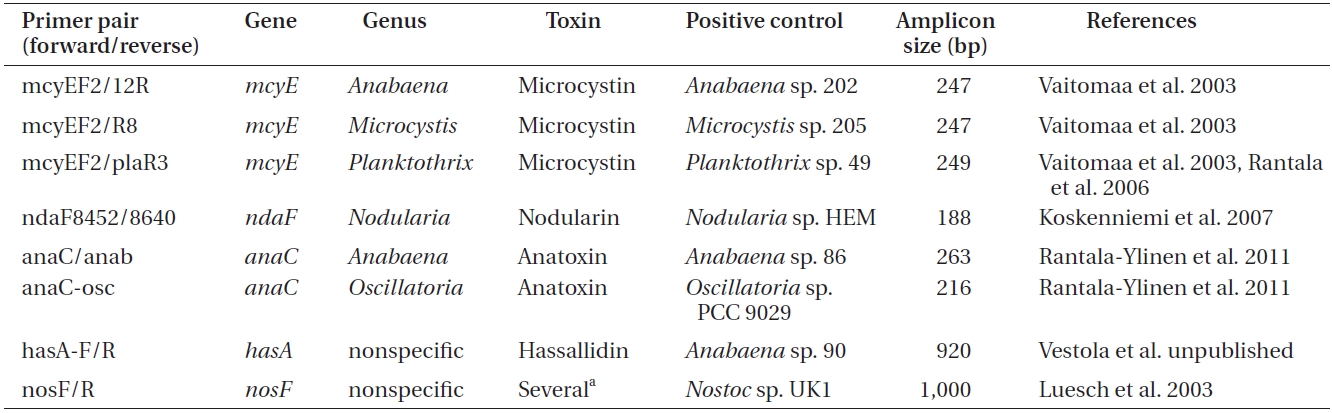
Primers used to amplify genes of the microcystin, nodularin, anatoxin and hassallidin, as well as methylproline-containing compounds biosynthetic gene clusters
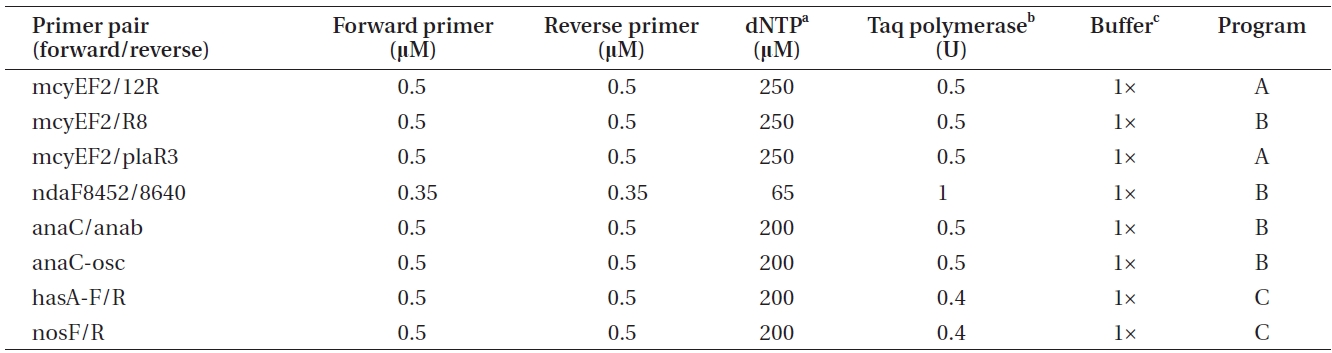
Polymerase chain reactions for the amplification of genes involved in toxin biosynthesis (Table 1) in a total volume of 20 μL
er of toxins, genomic DNA was amplified by conventional PCR using primers targeting a specific gene of the biosynthetic gene cluster of the toxin to be detected (Tables 1 & 2).
For each reaction (Table 2), 50 ng of genomic DNA was used as a template. As a positive control for each amplification, genomic DNA of the selected cyanobacteria was used (supplied by the University of Helsinki Cyanobacterial Culture Collection, UHCC) (Table 1), whereas sterile water was used as a negative control.
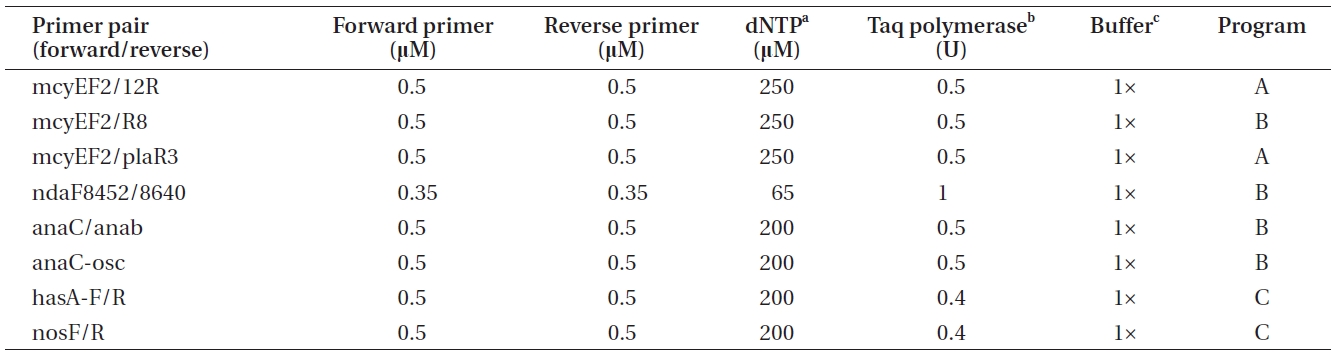
The PCR program A included the following phases: initial denaturation at 95℃ for 3 min, 30 cycles comprised of denaturation at 95℃ for 30 s, annealing at 58℃ for 30 s and annealing at 72℃ for 30 s, as well as final annealing phase at 72℃ for 5 min. The PCR program B was comprised of the same phases as program A, with the exception of the annealing phase being carried out at 60℃ for
30 s. PCR program C was 94℃ for 3 min, 36 cycles of 94℃ for 30 s, 52℃ for 30 s, and 72℃ for 1 min, as well as 72℃ for 10 min. Amplification products were checked by electrophoresis on 1.5 % (w/v) agarose gels followed by 0.10 μg mL-1 EtBr (Bio-Rad) staining and visualization on an UV transilluminator.
>
Evaluation of antifungal activity
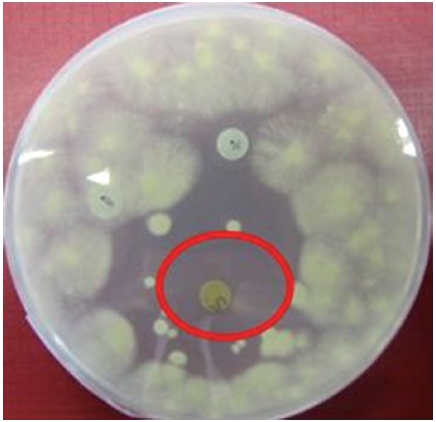
Extracted compounds of the ASN_M strain were examined for bioactivity against the yeast
Colonies of the
For each assay the following disks were prepared: a disk containing 730 μg of sample, another disk containing 10 mg of sample as well as a negative control disk containing 100 μL of methanol (LC-MS grade; Fischer Scientific) and a positive control disk containing 10 μg of nystatin.
ASN_M strain cell extracts were analysed by LC-MS using an Agilent 1100 Series LC/MSD Trap XCT Plus System (Agilent Technologies). Samples for analysis of anatoxin were separated with a Cogent Diamond Hydride column (100 A, 150 mm × 2 mm, particle size 4 μm; MicroSolv, Eatontown, NJ, USA), whereas samples for quantitation of microcystin where separated with a Luna C18(2) reverse phase column (100 , 150mm 2 mm, particle size 5 m; Phenomenex, Torrance, CA, USA) and samples for theanalysis of other bioactive compounds were separated with a Luna C8(2) reverse phase column (100 A, 150mm × 2 mm, particle size 5 m; Phenomenex). The injectionvolume of each sample was 10 μL. For anatoxin analysis, a commercial anatoxin-a standard (Enzo Life Sciences International, Farmingdale, NJ, USA) was used.
>
Morphological characterization and phylogenetic analysis of the 16S rRNA gene of ASN_M strain
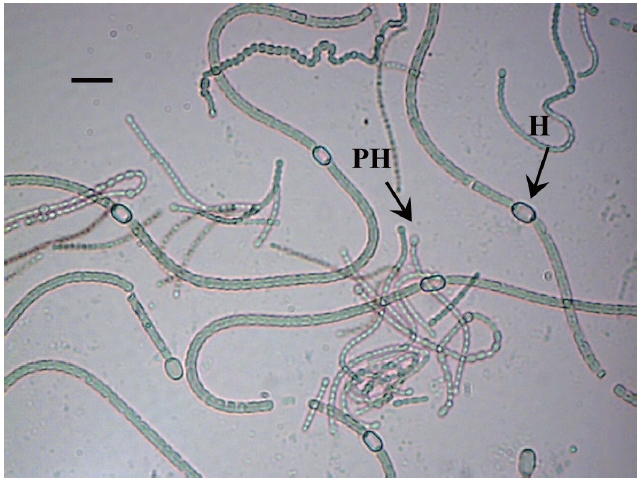
The isolate was identified as a filamentous cyanobacterial strain. The vegetative cells were spherical or slightly longer than broad (4-5 μm broad and 5.5-7 μm long), olive, heterocyts somewhat globose (4.5-7 μm broad and 4-8.5 μm long), akinetes oblong, many in a chain (5-6 μm broad and 6.5-11 μm long). Thallus gelatinous, adhering by under surface, dull olive in color (Fig. 2).
Based on the description of the morphology provided by Desikachary (1959) this cyanobacterium was identified as a strain of
An 837 bp portion of the 16S rRNA gene was successfully amplified from the PCR amplicon obtained from the ASN_M strain. The phylogenetic tree which was constructed using the neighbor-Joining method is shown in Fig. 1. Among available 16S rRNA sequences, the closest
relative to ASN_M strain was
>
Identification of potential of bioactive compounds producers by PCR
PCR results confirmed the bioactivity potential of the ASN_M strain. A 1,000 bp portion of the
>
Assaying Nostoc strain for antifungal activity
The methanolic extract of ASN_M strain had an antifungal effect on
>
Chemical identification of cyanobacterial toxins and bioactive compounds
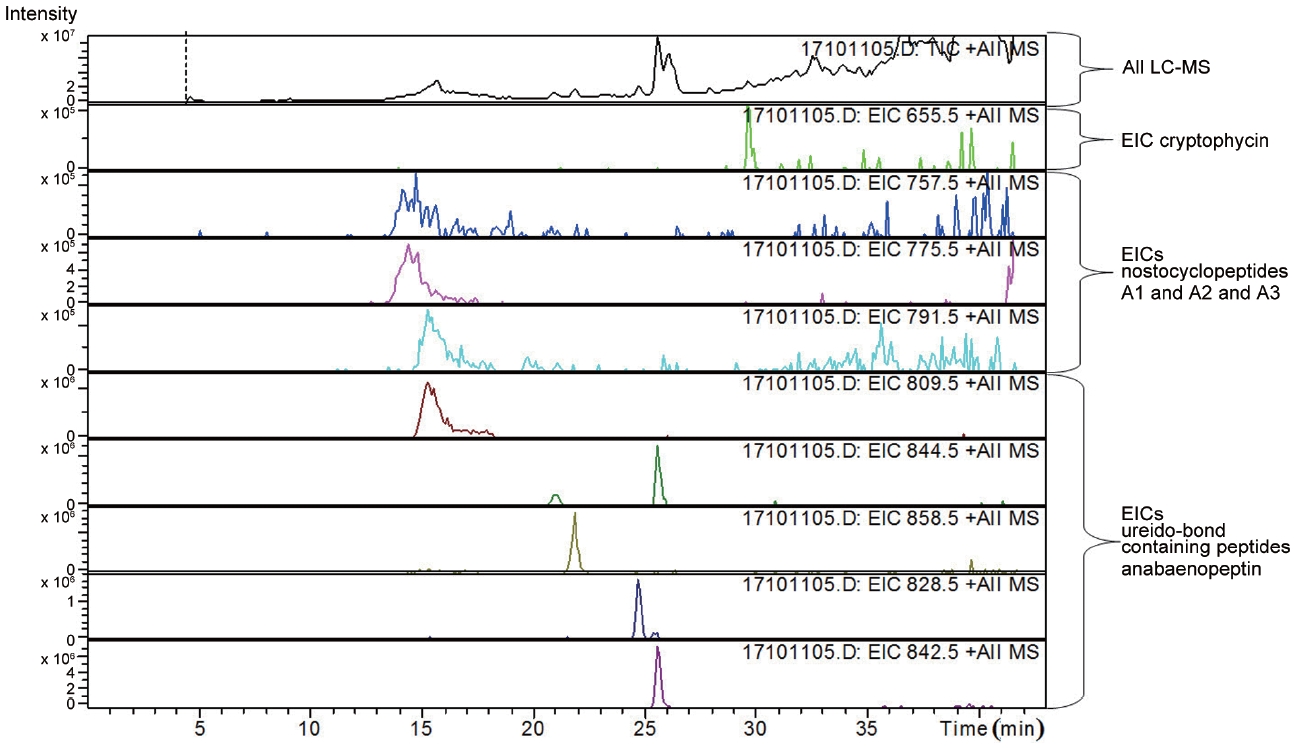
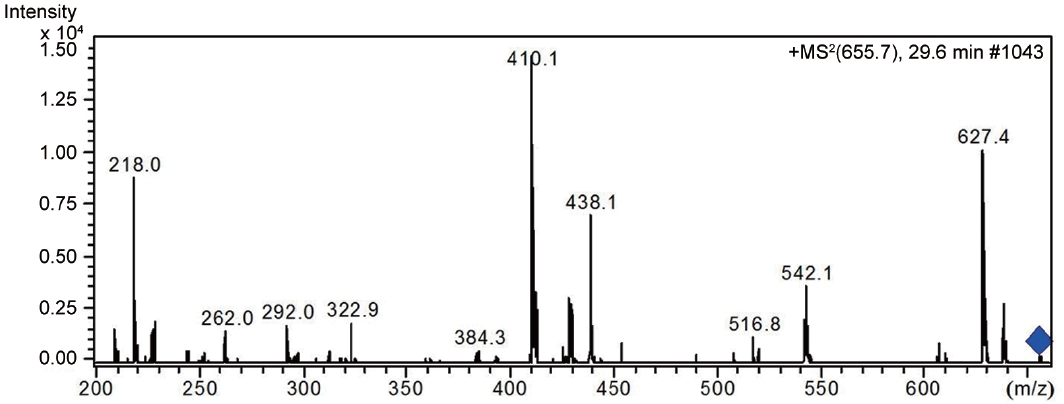
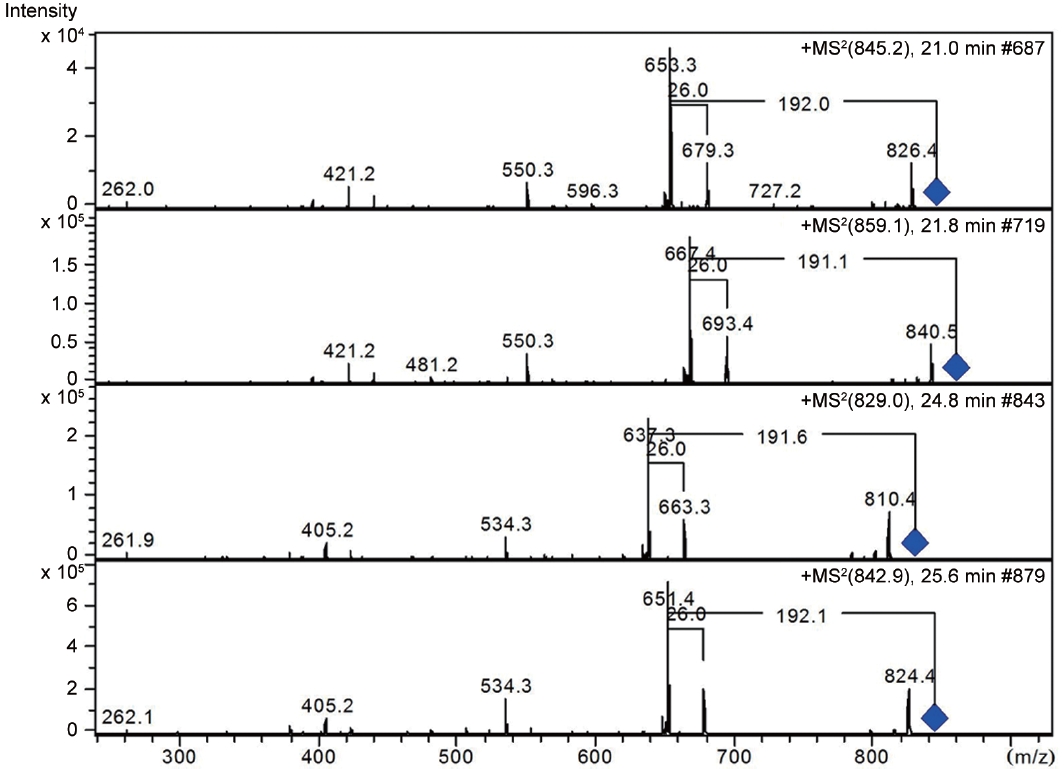
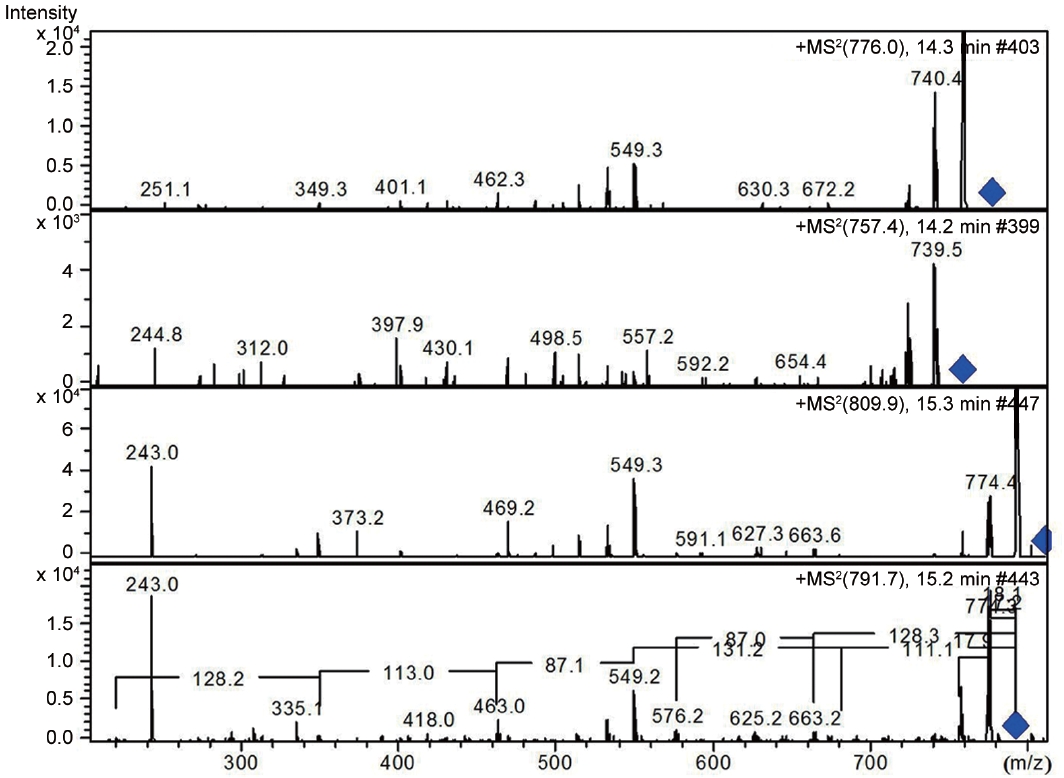
LC-MS analysis revealed the presence of three different classes of peptidic compounds (anabaenopeptins, cryptophycin, and nostocyclopeptides) from the ASN_M strain (Figs 4-7). No anatoxin-a, homoanatoxin-a, hassallidin or microcystin were detected in the ASN_M strain.
The classification of cyanobacteria has routinely relied on morphological characteristics which are not always trustworthy, as they may show variation depending on the culturing and environmental conditions, leading to misidentifications (Komarek and Anagnostidis 1989). In the present study, through a polyphasic approach, morphometric and genetic (16S rRNA) data were used to characterize the strain in a liquid suspension culture. The result of morphological characteristics of the ASN_M strain and genetic criteria showed congruency. Thereby, this cyanobacterium was named as
Cyanobacteria produce a great variety of secondary metabolites. These include cyanobacterial toxins but also compounds interesting for therapeutic use (Fewer et al. 2009).
Sixteen metabolites with bioactivities, such as anticancer, antibiotic, antifungal, and antiviral properties, have
already been found in seven
Molecular identification of bioactive compounds showed the amplification of
Interestingly, the presence of the peptidic compound nostocyclopeptide in the ASN_M strain was confirmed by LC/MS screening (Fig. 7). Nostocyclopeptides, a cyclic hexapeptide, showed weak cytotoxicity against KB (a human nasopharyngeal carcinoma) and LoVo (a human colorectal adenocarcinoma) cell lines and were devoid of antifungal and antibacterial activities (Golakoti et al. 2001, Becker et al. 2004, Jokela et al. 2010).
Anabaenopeptins have been reported to cause a relaxation of noradrenaline-induced contraction of aortic preparations in rats and anabeanopeptolides have been documented to cause inhibition of the serine protease chymotrypsin (Harada 2004). Moreover, Murakami et al. (2000) found that anabaenopeptins, ureido bound-containing cyclic peptides, have potent carboxypeptidase-A inhibitors (Fig. 6).
Besides nostocyclopeptides and anabaenopeptins, the obtained results showed the presence of the cryptophycin from the ASN_M strain (Fig. 5). The ability of the ASN_M strain to produce cryptophycins and the potency of its extract against
Although, the use of nostocyclopeptides and anabaenopeptins in medicine is not as wide spread as it once was, the use of cryptophycins in pharmacy, medicine, and biochemistry is well established.
Several types of semisynthetic analogue of cryptophycins with greater therapeutic efficiency and lower toxicity than cryptophycin are currently in clinical trials or preclinical trials or are undergoing further investigation.
Certainly, the exploitation of ASN_M strain as an organism with potential anti-fungal properties in the pharmaceutical industry has the potential to prevent many fungal diseases. Moreover, cryptophycins, by exerting antiproliferative and antimitotic activities by binding to the ends of the microtubules can be effective in treating many diseases, as the studies of Liang et al. (2005) showed excellent activity of cryptophycin against tumors implanted in mice.
Actively growing soil cyanobacterial mats may pose little risk as the majority of cyanotoxins are likely to be intracellular and not released into water. However, when mats die, as could be the case once a paddy filed is drained, there could be large- scale released of cyanotoxins (Smith et al. 2011).
These findings indicate that soil cyanobacteria represent a promising but still unexplored natural resource possessing many bioactive compounds useful for the pharmaceutical industry. However, the toxicity of theses soil cyanobacteria on soil microorganisms remains to be evaluated. This merits further and more detailed investigations.