Myosin is a hexameric protein consisting of two heavy chains (MHC; 200 kDa) and four light chains (MLC; 20 kDa). The light chains are classified into two classes: light chains released from heavy chains by alkali treatment (named alkali, essential light chains; MLC1 and MLC3) and light chains removable by 5,5´-dithio-bis-2-nitrobensoic acid (DTNB) treatment (referred to as regulatory light chains; MLC2) (Czosnek et al., 1982; Moutou et al., 2001). The second class (DTNB-removable) MLC2 family also exists as multiple isoforms exhibiting different tissue- and developmental stage-specific expression patterns, and the isoform MLC2f is predominantly expressed in fast skeletal muscles (Chu et al., 2011). Although several previous studies have reported differential expression patterns of fish myosin-related genes modulated by developmental stage, hormone treatment, and water temperature (Moutou et al., 2001; Temple et al., 2001; Hall et al., 2003), the molecular mechanism of fish mlc2f gene expression remains poorly understood compared to the substantial information available for mammalian and avian orthologs.
To date, comparative analyses on coding sequences of teleost mlc2f genes have shown that teleostean orthologs share a considerable degree of structural homology at both the nucleotide and amino acid levels (Mugue et al., 2005; Mugue and Ozernyuk, 2006). However, at the genomic level, the fish mlc2f gene has been underexploited, and more importantly, the structural and functional characteristics of the teleost mlc2f promoter have not yet been comprehensively studied. Although transgenic studies on zebrafish (Danio rerio: Cypriniformes) have highlighted that the zebrafish mlc2f promoter could drive the strong, muscle-specific expression of foreign proteins (Xu et al., 1999; Wan et al., 2002; Ju et al., 2003), the functional and structural evaluation of mlc2f promoters from other fish species has been achieved in relatively few studies (Krasnov et al., 2003; Funkenstein et al., 2007).
The Javanese ricefish (Oryzias javanicus: Beloniformes) is an emerging candidate as a model fish system. Owing to its excellent ability to acclimate to a wide range of salinities, a great deal of attention has focused on this species as a test model organism, not only to study molecular mechanisms and diversification of seawater adaptation in teleostean groups, but also to perform ecotoxicological in vivo biomarker assays for both freshwater and marine ecosystems (Koyama et al., 2008). In addition, this small, egg-laying fish species possesses a readily transparent body color throughout its life cycle (Song et al., 2010), which is highly desirable for many experiments requiring the in vivo presentation of fluorescent protein reporter(s).
The objectives of this study were to isolate and characterize the genetic determinant of the mlc2f gene at both the mRNA and genomic levels in Javanese ricefish and to examine the potential utility of the mlc2f promoter in driving the muscleenriched heterologous expression of the fluorescent reporter in microinjected embryos.
A cDNA library was constructed with pooled tissues of Javanese ricefish using a Lambda Zap XR cDNA Synthesis Kit (Stratagene, La Jolla, CA, USA). From our expressed sequence tag (EST) database generated from the Javanese ricefish cDNA library (unpublished data), EST clones (n = 6) showing high homology to known vertebrate MLC2f were assembled into a contig using Sequencher software version 4.2 (Gene Codes, Ann Arbor, MI, USA). Vectorette polymerase chain reaction (PCR) was performed using the excised stock of the cDNA library to obtain the 5´-untranslated region (UTR) and 3´-UTR. The continuous segment of the full-length mlc2f cDNA was isolated again using reverse transcription (RT)-PCR of Javanese ricefish whole body total RNA, and the sequence was confirmed. Based on the cDNA sequence, a genomic fragment of mlc2f spanning from 5´-UTR to 3´-UTR was PCR-isolated using a pair of PCR primers, OJMLC2g FW (5´-GATATACGTAAAACGCGGCT-3´) and OJMLC2g RV (5´-GACTTTACAGGATCTTTTTATTG-3´). The PCR product was directly sequenced in both directions using a conventional primer walking method. The open reading frame (ORF) was predicted using the ORF finder in NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), and the amino acid sequence was also deduced from the ORF sequence using the same tool. The deduced amino acid sequence of Javanese ricefish MLC2f was aligned with other teleostean MLC2fs retrieved from public databases using ClustalW (http://align.genome.jp). The Ex- PASy ProtParam tool (http://web.expasy.org/protparam/) was used to estimate the theoretical molecular weight (kDa) and isoelectric point (pI) of the MLC2f amino acid sequence. The genomic organization of the Javanese ricefish mlc2f gene was compared with those from other teleost orthologs, which were deciphered from Ensembl Genome Browser (http://www.ensembl.org/index.html). Identification of repetitive sequences in introns of mlc2fs was conducted using the Unipro UGENE software (http://ugene.unipro.ru/).
To obtain the 5´-upstream region of mlc2f, genome walking was performed using the Universal GenomeWalker Kit (Clontech Laboratories Inc., Mountain View, CA, USA). All procedures for constructing genome walking libraries with DraI-, EcoRV-, PvuII-, or StuI-digested O. javanicus genomic DNA and two successive PCR amplifications using the gene-specific, reverse primers (ODMLC2 GW1, 5´-CATCTTGAGAAGCTGTGAGTGGAGCT- 3´ and ODMLC2 GW2, 5´-TGGCCGATCAAGAAAGAGAAGACAG- 3´) were performed according to the manufacturer’s recommendations. Amplified product was purified using the QIAEX II Gel Extraction Kit (Qiagen, Hilden, Germany), cloned into the pGEM T-easy vector (Promega, Madison, WI, USA) according to the manufacturer’s instruction and sequenced. Based on the sequence assembled in contig, the continuous genomic sequence spanning from promoter to the 3´-UTR of the structural gene was PCR-isolated using OJMLC2p 1F (5´-GACTGCATCATTTCCTTAAGC- 3´) and OJMLC2g RV (see above) and then sequenced in both directions. Potential transcription factor (TF) binding motifs in the 5´-flanking upstream region of the Javanese ricefish mlc2f gene were predicted using TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess?RQ=WELCOME) and TF search (http://www.cbrc.jp/research/db/TFSEARCH.html).
Quantitative real-time RT-PCR was performed to examine the basal expression levels of mlc2f mRNAs in Javanese ricefish tissues and embryos. Tissue samples used for the quantitative RT-PCR assay were brain, eye, fin, gill, heart, intestine, kidney, liver, skeletal muscle, and spleen, all of which were obtained from 12 normally grown adults under conditions of 15 ppt salinity. Embryonic samples (approximately 100-300 embryos) were obtained from (I) just fertilized, (II) early cleavage (i.e., four- to eight-celled), (III) morula, (IV) blastula, (V) gastrula, (VI) neurula, (VII) four-somite stage, (VIII) heart beating stage, (IX) retinal pigmentation stage, (X) gill blood vessel formation stage, (XI) visceral blood vessel formation stage, (XII) spleen development stage, and (XIII) just-hatched. Details for developmental stages of this species can be found in Song et al. (2010). A total RNA sample was prepared using an RNeasy Mini kit (Qiagen) according to the manufacturer’s instruction, including a DNase I treatment step. Reverse transcription to cDNA was performed using the Omniscript Reverse Transcription Kit (Qiagen) according to the manufacturer’s manual. As a normalization control, the Javanese ricefish 18S rRNA primer was included in the RT-reaction (unpublished data). A diluted cDNA template (fourfold for mlc2f and 40-fold for 18S rRNA) was subjected to real-time PCR cycling using 2× iQ SYBR Supermix (Bio-Rad, Hercules, CA, USA). The reaction was run using the iCycler iQ Real-Time Detection System (Bio-Rad). Oligonucleotide primers for qPCR of mlc2f mRNA were qOJMLC2 1F (5´-AGGTTCTCCAAGGACGAGAT-3´) and qOJMLC2 1R (5´-TCTACAGACCATCTCGTGTG-3´), whereas qOJ18S RNA 1F (5´-TCCAGCTCCAATAGCGTATC-3´) and qOJ18S RNA 1R (5´-AGAACCGGAGTCCTATTCCA-3´) were used for 18S rRNA. Based on the standard curves, the PCR efficiency was confirmed to be higher than at least 93% in triplicate assays for both genes. The relative expression level of each sample was normalized against its own 18S rRNA level as described by Kubista et al. (2006). Differences in means based on triplicate assays were assessed using analysis of variance (ANOVA) followed by Duncan’s multiple range test at a level of P = 0.05 using SPSS software version 10.1.3 (SPSS Inc., Chicago, IL, USA).
To construct a red fluorescent protein (RFP) reporter vector under control by the Javanese ricefish mlc2f promoter, a 2.5- kb promoter region was isolated by PCR using SalI-OJMLC2p 2F (5´-AATGTCGACGACTGCATCATTTCCTTAAGC-3´) and AgeI-OJMLC2p 1R (5´-AATACCGGTCTTGAGAAGC TGTGAGTGGAGCT-3´) primers, of which the 5´-ends were designed to possess SalI and AgeI sites, respectively, to facilitate the subsequent cloning step. The PCR-amplified product was TA-cloned and spliced from the T-easy vector (Promega) by digestion with the two restriction enzymes (New England Biolabs, Ipswich, MA, USA). Digested fragment was unidirectionally spliced into just upstream of the ATG initiation codon of the RFP gene (rfp) in a pDsRed2-1 plasmid vector (Clontech Laboratories Inc.). Configuration and the correct sequence of the expression cassette were confirmed using sequencing analysis, and the construct was named pOJmlc2RFP (6.6 kb).
The pOJmlc2RFP construct was linearized by digestion with ApaLI (recognition sequence, GTGCAC) and gel-purified. Purified plasmid resuspended in an injection buffer (10 mM Tris-Cl, 0.1 mM EDTA, pH 8.0) was microinjected into a one-celled embryo using a Narishige MMN-330 micromanipulator (Narishige Scientific Instrument Lab, Tokyo, Japan). The DNA concentration for microinjection was 50 μg/mL, and the estimated amount of DNA delivered to each embryo was 10 pg on average. Microinjected embryos were placed in an incubator at 25℃ latipes until hatching. The salinity level was 10 ppt throughout embryonic development and larval growth. The appearance of RFP signals in microinjected embryos was monitored using an AZ100 epifluorescence microscope (Nikon Co., Tokyo, Japan) and photographed using the NIS-Elements Microscope Imaging Software implemented using the microscope system.
The O. javanicus mlc2f cDNA consisted of a 74-bp 5´-UTR, a 510-bp ORF encoding a 170-aa polypeptide, and a 204-bp 3´-UTR including a 17-bp poly(A)+ tail. The polyadenylation signal (AATAAA) occurred at 22 bp before the poly(A)+ tail (GenBank accession no. JQ905605). Based on the multiple sequence alignment, the deduced polypeptide sequence of O. javanicus MLC2f (19.0 kDa; pI = 4.69) also shares considerable homology with its vertebrate orthologs: the overall sequence identity between O. javanicus MLC2f with teleostean orthologs ranged from 87% to 98% (alignment not shown). The O. javanicus MLC2f conserved the helix-loop-helix (EF-hand) typical of vertebrate MLC2 proteins. A putative Ca2+-binding domain (IDQNRDGIISKDDLR) and potential phosphorylation site (Ser18) were predicted to be similar to most MLC2f proteins from teleost species (see JQ905605).
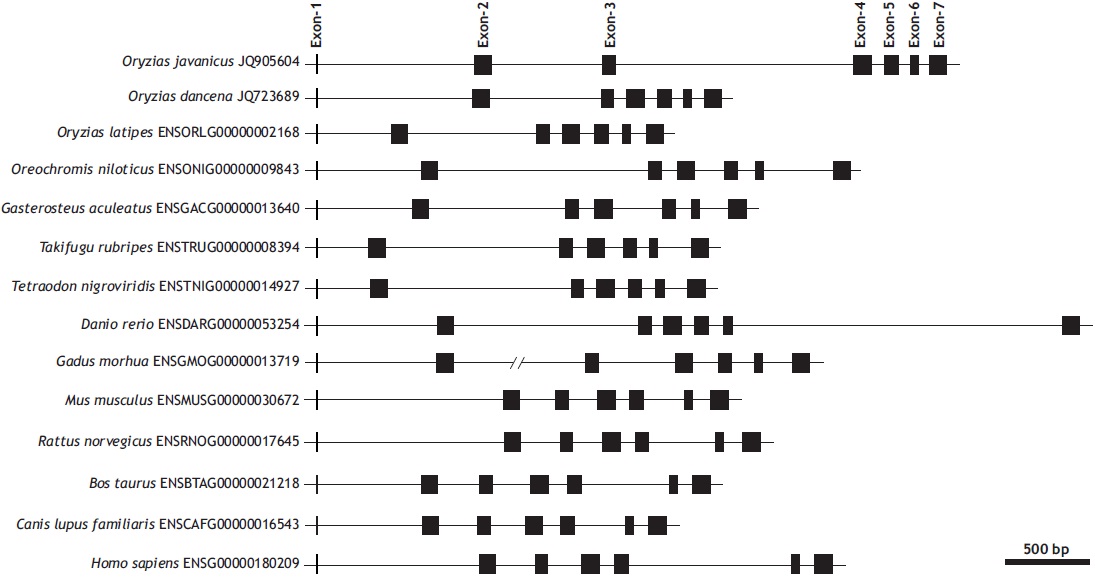
The genomic gene of O. javanicus mlc2f represented seven exons interrupted by six introns (935, 654, 1,419, 80, 69, and 66 bp, respectively, for introns 1-6), with a conserved GT/AG splicing rule in each exon-intron junction site (GenBank accession no. JQ905604). The first exon codes only one amino acid (Met encoded by the initiation codon ATG), and the remaining six exons code 33, 26, 35, 26, 16, and 33 amino acids, respectively. The seven-exon structure of mlc2f is conserved in vertebrate orthologs, in which each mlc2f gene codes Met1 in only its exon 1. The length of each coding region is highly similar regardless of the organism, but those of intronic and untranslated sequences are quite variable (Fig. 1).
Of the six introns in O. javanicus mlc2f, the two largest
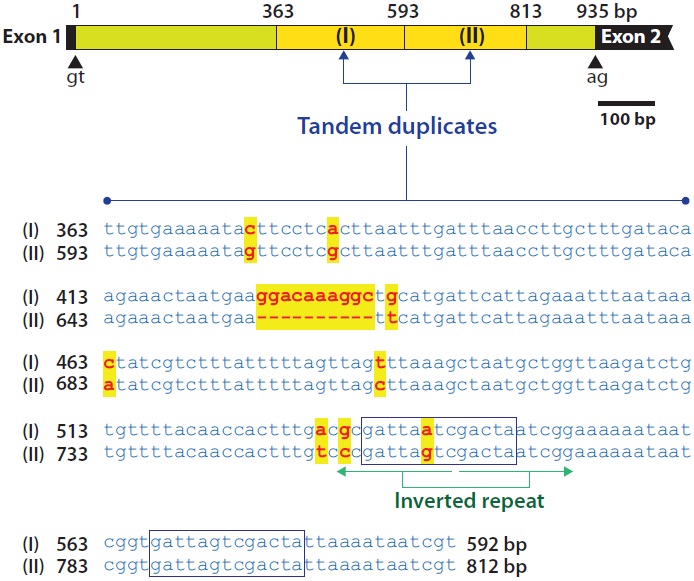
introns (935-bp intron 1 and 1,419-bp intron 3) were highly A + T-rich, (69% and 68%, respectively, for introns 1 and 3), whereas the remaining exons exhibited A + T percentages ranging from 47.9% to 61.3%. In addition, introns 1 and 3 contained duplicated and/or repetitive sequences. In intron 1, a 230-bp region beginning 363 bp from the 5´-end of the intron (TTGTGAAAAT…AAATAATCGT) was duplicated in a tandem fashion. The second copy resulting from duplication was 220 bp in length due to a deletion of a 10-bp sequence (GGACAAAGGC) of the first copy. With the exception of this deletion, nucleotide substitutions between these duplicated copies were only found in eight positions (96% sequence identity). In the duplicated region, a 13-bp sequence (GATTARTCGACTA) containing a palindrome sequence (TARTCGACTA) occurred twice (Fig. 2).
The repetitive pattern in intron 3 was much more complex. A 155-bp unit sequence (TT[T/A]AGGC…CATTTTT) was tandemly repeated six times, and approximately half of the 155-bp unit was additionally repeated once, consequently forming a 1-kb internal tandem repetitive region. Moreover, the 155-bp sequence of the internal repetitive region included several of the same subunit sequences as those found in either 5´-end (Fig. 3).
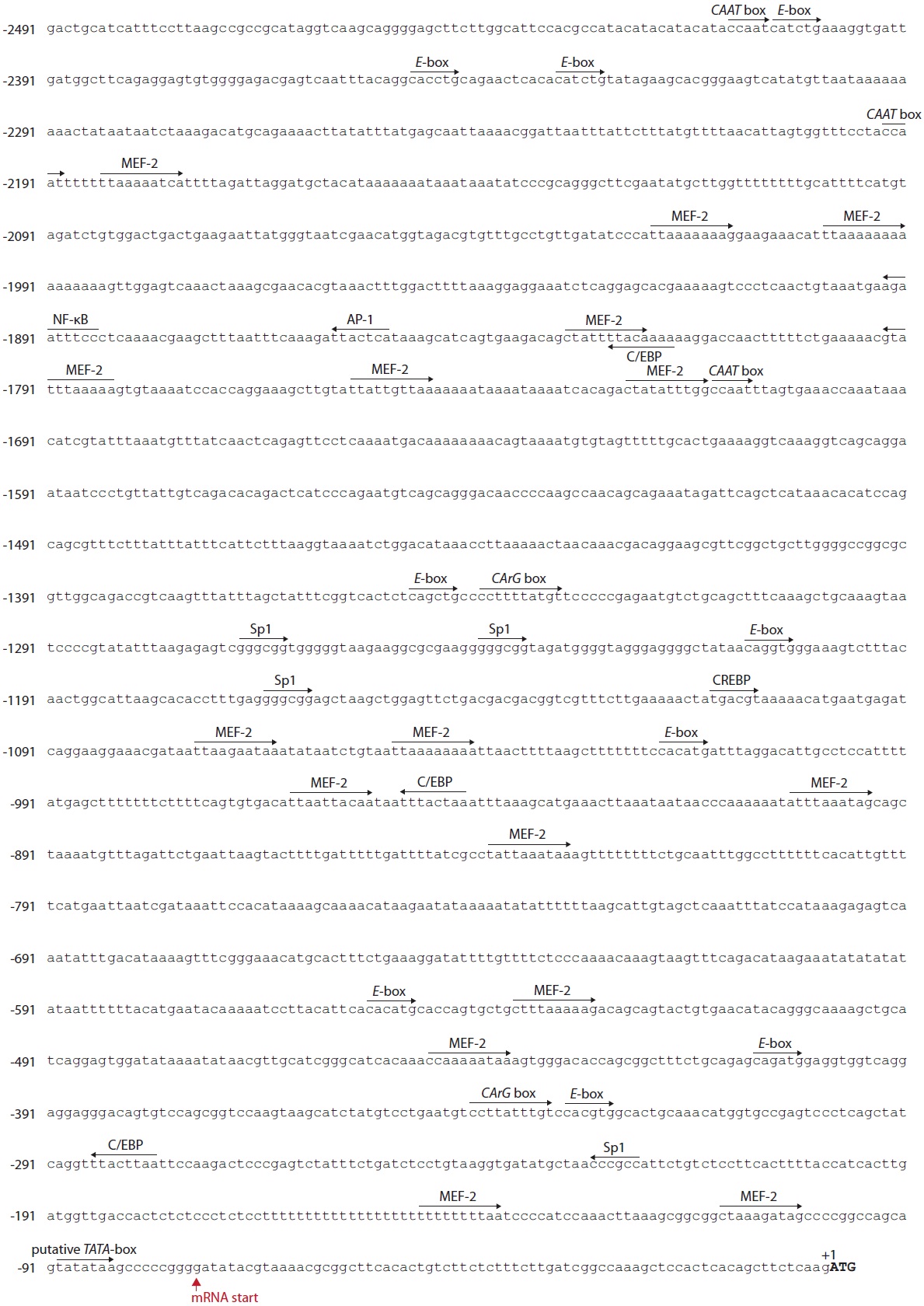
Bioinformatic mining of TF-binding motifs from the 5´-flanking region (a 2,491-bp upstream region from the ATG initiation codon) showed that the O. javanicus mlc2f promoter possessed potential sites bound by various TFs (Fig. 4). A putative TATA box was found at -90 bp from the ATG translation start site (corresponding to -16 bp from the transcription start site) in the less common form, TATATAA. A poly(T)23?26 was found at -76 bp from the TATA box with a length polymorphism among PCR clones (data not shown). Three copies of a CAAT box (consensus sequence, CCAAT) were found in the distal promoter regions, and two CArG boxes (CCW- 6GG) were predicted in a noncanonical form at -342 (CCTTATTTGt) and -1,341 bp (CCTTTTATGt), respectively, from the ATG codon. Several important TF-binding sites related to muscle-enriched and/or targeted expression were predicted. Multiple copies (n = 16) of myocyte enhancer factor-2 (MEF- 2) binding sites were distributed throughout the 5´-flanking region of the O. javanicus mlc2f gene with 90% sequence identity to the consensus form ([Y=C/T]TAW4TAR) or YTWWAAATAR. A total of nine copies of an E-box (CANNTG), which is potentially targeted by MyoD and/or myogenin, was identified in both the proximal and distal regions. In addition to these essential and/or muscle-related elements, several other TF-binding sites were also predicted: activator protein-1 (AP-1), Sp1 transcription factor, CCAAT-enhancer binding protein (C/EBP), cyclic AMP response binding protein (CREBP), and nuclear factor kappa B (NF-κB; GGRNNYYCC).
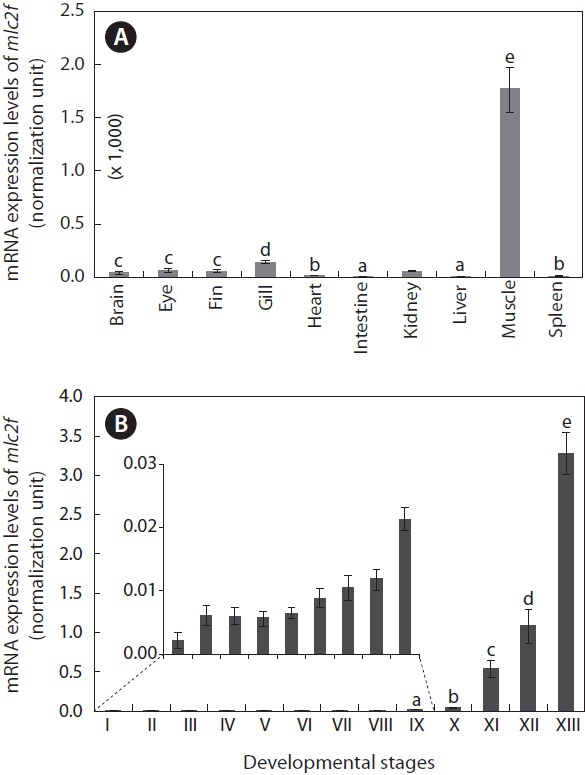
The mlc2f transcripts were highly enriched in skeletal muscles (Fig. 5A). For other tissues, the mlc2f transcripts were detectable, but very weakly, as assessed by real-time RT-PCR (P < 0.05). In addition to the skeletal muscle, which exhibited the highest expression level, relatively high expression was observed in the gill (P < 0.05), although at one-tenth the level compared to that in skeletal muscle. The intestine and liver exhibited the lowest levels of mlc2f expression (P < 0.05). The mRNA expression of mlc2f was also significantly modulated during the embryonic development of O. javanicus (Fig. 5B). Upon fertilization, the mRNA level of mlc2f was slightly elevated, but no further increase occurred during early development of embryos until the gastrula stage (P > 0.05). From the neurula stage to the retinal pigmentation stage, the mlc2f
[Fig. 5.] Tissue distribution (A) and embryonic expression (B) of Oryzias javanicus mlc2f mRNAs as determined by quantitative reverse transcription-PCR based on the normalization against 18S rRNA. Means with different letters are not significantly different based on ANOVA followed by Duncan’s multiple range tests at P < 0.05. In (B), the mlc2f expression in early developmental stages was very low, and thus the expression levels only from stages IX to XIII were subjected to statistical evaluation. Developmental stages are (I) just fertilized, (II) early cleavage (i.e., four to eight-celled), (III) morula, (IV) blastula, (V) gastrula, (VI) neurula, (VII) four-somite stage, (VIII) heart beating stage, (IX) retinal pigmentation stage, (X) gill blood vessel formation stage, (XI) visceral blood vessel formation stage, (XII) spleen development stage, and (XIII) just-hatched.
mRNA level gradually increased. Subsequently, transcription of the mlc2f gene was markedly activated as development proceeded. The mlc2f mRNA level in hatched larvae was more than 85-fold relative to that observed in the gill vessel formation stage (P < 0.05).
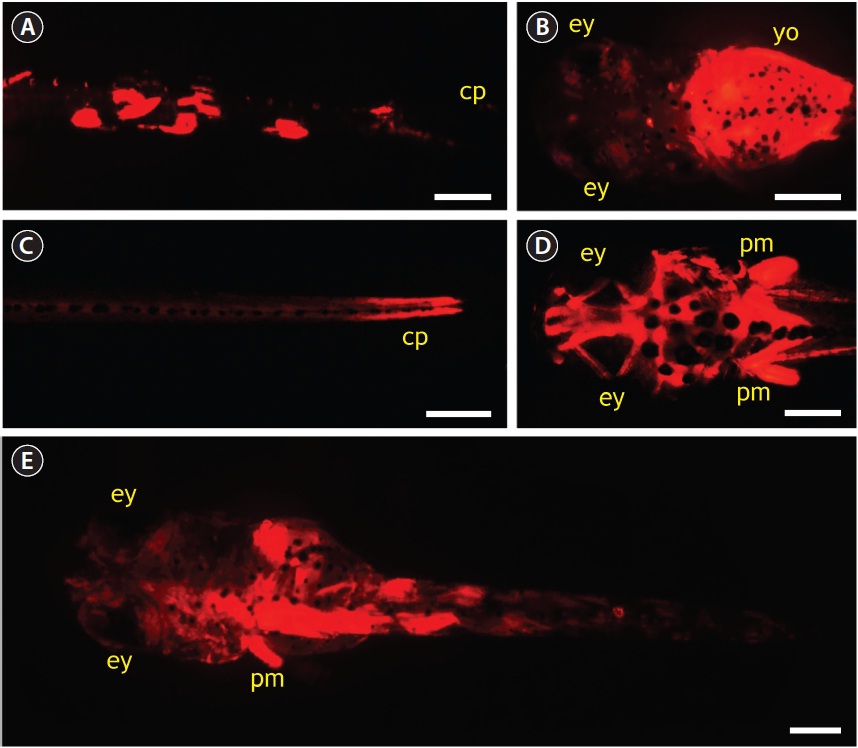
Microinjection using pOJmlc2RFP (6.6 kb) was performed to evaluate the potential utility of O. javanicus mlc2f promoter in driving the skeletal muscle-specific expression of a downstream foreign gene. In total, 652 one-celled embryos were microinjected during three trials, and the average hatching success was 36%, which was about 40% of that observed in the non-injected control. None of the embryos injected with pOJmlc2RFP showed RFP signals during early developmental stages. Only after embryos reached the blood circulation stage did a small portion of embryos (about 5% of the surviving embryos) begin to express RFP signals (photograph not shown). Afterward, about 25% of microinjected embryos began to display RFP expression, and the RFP signals gradually intensified as development progressed. However, the expression of RFP was mosaic in nature; that is, not all skeletal muscles expressed the RFP, consequently leading to variable patterns of RFP distribution among resultant hatched larvae. About half of the RFP-positive larvae showed a mosaic distribution of RFP signals, mainly in dorsal and/or peduncle muscles (Fig. 6A). A considerable portion of larvae (up to 20%) showed yolk-dominant expression of RFP (Fig. 6B). The RFP expression in yolk disappeared after yolk-sac absorption. Approximately 30% of RFP-positive larvae showed strong RFP expression in caudal peduncle and head muscles (Fig. 6C and 6D). The strong RFP expression in head muscles was often accompanied by significant RFP signals in pectoral fin muscles. Several larvae maintained persistent RFP signals during their early life phase after the first feeding (Fig. 6E).
Javanese ricefish MLC2f revealed conserved structural features that are typical of the EF-hand superfamily composed of the helix-loop-helix motif structure found in all other MLC2f orthologs from vertebrates, including teleosts. The O. javanicus MLC2f also shares a potential phosphorylation site at Ser18 that is conserved in all vertebrate orthologs. Phosphorylation of MLC in skeletal muscles of mammals by MLC kinase is Ca2+-dependent (Ikebe et al., 1998; Fujita et al., 1999), and the phosphorylation is closely related to myosin ATPase activity and muscle contraction (Tang and Gunst, 2004). Thus, fish MLC2fs, including the present O. javanicus MLC2f, are likely to be phosphorylated in this Ser residue (see also Sweeney, 1995).
At the genomic level, the lengths of introns are highly variable among vertebrate species despite strong similarity in the lengths of each coding exon. However, the total length of noncoding regions is not directly proportional to the genome sizes of these organisms. Javanese ricefish mlc2f shares a conserved exon-intron organization with its vertebrate orthologs. Our molecular phylogenic analysis inferred that the Javanese ricefish mlc2f gene belongs to the teleost mlc2f a group characterized by the seven-exon structure (data not shown). However, unlike other teleostean orthologs, Javanese ricefish mlc2f exhibits multiple tandem duplication events in its introns. A closer look at the intronic sequence indicates that the O. javanicus mlc2f gene contains tandem repeats in introns 1 and 3. In intron 1, a relatively large fragment (230-bp fragment) is tandemly duplicated, causing the following 220-bp fragment to have a sequence identity higher than 90%. In contrast, intron 3 contained 6.5 tandem repeats of a 155-bp unit
sequence with complex tandem and inverted repeats of shorter unit sequences in both flanking regions of the internal tandem repeats. The fish myosin gene family is known to have undergone gene duplication and divergence into diverse paralogs via fish-specific whole genome duplication events during their evolutionary history (Mugue and Ozernyuk, 2006). However, such tandem duplication(s) within intron(s) has not been reported in vertebrate mlc2f genes. A bioinformatic search showed that the tandem-repetitive pattern seen in the intron of the O. javanicus mlc2f was not clearly observable in other teleostean orthologs, including those from other Oryzias species (e.g., O. latipes and O. dancena). These results suggest that the “intra-intronic” duplication events may be Javanese ricefish-specific rather than Oryzias-specific. However, we found a potential duplication of an intronic sequence in the human mlc2f gene (NC_000016.9), although the pattern in the human ortholog differs from that in O. javanicus mlc2f. In the human ortholog, a 140-bp region in intron 1 is duplicated as a 141-bp copy in intron 5 in the same orientation. However, the nucleotide sequence identity between the two duplicated copies in human mlc2f is only 89%. Also, our extension of the bioinformatic analysis to other primate mlc2f orthologs indicates that mlc2f genes from many primate species also possess the duplication of an intronic fragment between intron 1 and intron 5. Furthermore, the duplicated intronic sequences are conserved among these primate orthologs, suggesting that the “inter-intronic” duplication is primate-specific. However, we have not yet clarified the biological or evolutionary significance of the tandem repetitive pattern in the Javanese ricefish, although this issue will be explored further. Even though we have not yet found individual variations in the length polymorphism of this repetitive region using several Javanese ricefish stocks, further examination of various strains or local populations may be needed to obtain deeper insight into the patterns of the intronic duplications in this fish species.
A bioinformatic analysis of a 2.5-kb 5´-flanking region of O. javanicus mlc2f showed a putative TATA box in a TATATAA sequence rather than the more commonly found core sequence, TATAAA. Our search of other vertebrate orthologs indicated that all vertebrate orthologs also have the same TATATAA sequence. Two other essential boxes, the CAAT (perfectly matched to the consensus sequence) and CArG (imperfect form with only 90% identity to the consensus sequence) boxes, were also predicted in multiple copies; however, their functional roles in the regulation of fish mlc2f genes have not been clearly characterized experimentally. In addition to these general boxes, multiple copies of an E-box and an MEF-2 site, both of which are known to be responsible for muscle-specific or enriched gene expression, were identified. The E-boxes (CANNTG) are binding sites recogconnized by myogenic basic helix-loop-helix transcription factors (e.g., MyoD and myogenin) and are commonly found in many muscle-specific genes, including different myosin light chain genes (Kobiyama et al., 1998; Watabe, 1999; Du et al., 2003). The efficient onset of skeletal myocyte differentiation requires the binding of MyoD family proteins, as heterodimers with E2A, to this common DNA sequence element (E-box) (Galloway et al., 2006). The MEF-2 functions as a critical, central regulator for myogenesis in both cardiac and skeletal muscles. The consensus sequence YTAW4TAR is targeted by homo- and heterodimers of MEF-2 (Black and Olson, 1998; Funkenstein et al., 2007). Previous studies have also shown that almost all cardiac and skeletal muscle-specific genes may possess multiple copies of MEF-2 binding sites in their promoters (Esser et al., 1999; Xu et al., 1999). The 2.5-kb O. javanicus mlc2f promoter isolated in this study displays multiple copies of MEF-2 binding sites with 90% sequence identity to the consensus sequence. Comparison of proximal and distal promoter sequences of vertebrate mlc2f genes indicated that the number of copies of MEF-2 sites differed depending on the species (data not shown), suggesting, at least in part, that the MEF-2-mediated regulation of mlc2f genes at the transcriptional level may be diversified among species. Nevertheless, functional typing of each MEF-2 site would be needed, as a previous study reported that even a short proximal promoter would be sufficient to display sufficient activity of mlc2f, as judged by heterologous expression assays (Ju et al., 2003). The O. javanicus mlc2f promoter also represents several binding sites for different TFs, including CREBP, Sp1, AP-1, and NF-κB, which are closely related to stress and immune responses. These TF-binding sites might also be observable in other fish mlc2f promoters in our bioinformatic analysis. However, the functional involvement of these TFs in the regulation of fish mlc2f genes has yet not been determined. In contrast, previous studies on the mammalian mlc2f have reported that downregulation of AP-1, Sp-1, and NF-κB should be a prerequisite for the onset of myocyte differentiation (Lehtinen et al., 1996; Sachdev et al., 2003).
The mRNA expression of O. javanicus mlc2f was highly predominant in skeletal muscles, whereas the expression levels in other tissues were minor or even negligible. Skeletal muscle-enriched expression of mlc2f has already been reported in other fish species (Xu et al., 1999; Ozernyuk et al., 2004). During embryogenesis of O. javanicus, mlc2f expression was differentially modulated. The expression of mlc2f transcripts during embryogenesis was not significant until the retinal pigmentation stage, and afterward, its expression sharply increased until hatching. Different isoforms of myosin light chains are known to represent differential expression patterns during embryonic development, and the mlc2f isoform is often detectable from the late somite stages in fish embryos (Galloway et al., 2006). The present findings for O. javanicus mlc2f broadly resembled those previously described for other fish species, although the point at which mlc2f transcripts sharply increase (i.e., gill blood vessel formation stage) in this species was slightly lagged compared to those previously observed in other species (Xu et al., 2000; Ju et al., 2003; Galloway et al., 2006). Hence, further examination is needed, particularly comparative assays using both whole-mount in situ hybridization and quantitative RT-PCR.
RFP expression in the pOJmlc2RFP-microinjected group was clearly visible, mainly beginning at the gill blood vessel formation stage, and the intensity of the RFP signals became stronger as development progressed. Although this expression pattern is in broad agreement with that of endogenous mlc2f mRNA expression as determined by the quantitative RT-PCR assay, the timing of the onset is slightly delayed. This temporal delay is likely related to the lag period for the accumulation of RFP to a detectable level. However, further validation of the onset expression and spatial distribution of RFP signals should be made using stable transgenic germlines, as the mosaic distribution of injected transgene may cause an even more extended lag time for the accumulation of sufficient RFP to a detectable level (see also Ju et al., 2003; Krasnov et al., 2003). As expected, RFP-positive larvae exhibited a mosaic distribution of RFP in their skeletal muscles due to the mosaic nature of the transgene. However, the main expression sites of RFP were dorsal skeletal muscles, caudal peduncle muscles, head muscles, and pectoral fin-attaching muscles, which was in agreement with the expected patterns of the endogenous mlc2f gene. Similar results were also observed in zebrafish mlc2f promoter-driven transgenic fishes (Ju et al., 2003; Gabillard et al., 2010). In the present study, microinjection of pOJmlc2RFP resulted in several larvae showing ectopic RFP expression (e.g., RFP signals in the yolk sac and eye lens). Similar findings were also observed in rainbow trout Oncorhynchus mykiss embryos microinjected with muscle-specific promoter-driven reporter genes; the authors proposed that the yolk syncytial layer endowed with actin-containing, contractile microfilaments exhibited a high capacity for transgene expression (Krasnov et al., 2003; Carvalho and Heisenberg, 2010). In general, ectopic expression often disappears in subsequent generations of stable transgenic germlines (Ju et al., 2003; Cho et al., 2011). For this reason, establishment of stable transgenic germlines is needed to visualize both temporal and spatial expression of mlc2f in a more detailed and accurate manner.
In summary, the genomic organization and basal expression of mlc2f were characterized in Javanese ricefish, and the potential utility of the mlc2f promoter in driving musclespecific heterologous expression was examined. The Javanese ricefish mlc2f showed conserved features of fish mlc2fs with respect to exon-intron organization and secondary polypeptide structure. However, the Javanese ricefish mlc2f also possessed species-specific duplication events in its introns. The mRNA expression patterns of mlc2f in adult tissues and developing embryos were in agreement with those of mlc2fs in other teleosts. Microinjection of mlc2f promoter-driven reporter construct successfully exhibited the heterologous expression of the fluorescent reporter, primarily in muscular areas of embryos and resultant larvae. However, due to the mosaic nature of introduced transgenic DNA, stable transgenic lines may be required to elucidate the myogenesis of this species in detail.






![Internal tandem repeats in the intron 3 of Oryzias javanicus mlc2f gene. Six and half copies of a 155-bp unit (TT[T/A]AGGC…CATTTTT) are repeated in a tandem fashion.](http://oak.go.kr/repository/journal/12012/E1HKAL_2012_v15n4_325_f003.jpg)