Alginate, a polysaccharide that naturally contains carboxyl groups in each constituent residue (Ikeda et al., 2000), has been used in the food, pharmaceutical and medical industries (McNeely and Pettitt, 1973; McCleary et al., 1983). The industrial usage of alginate is based on three major characteristics. The first ability of alginate is to thicken when dissolved in water. The second is to form gels when a calcium salt is added to a solution of sodium alginate in water. The third is the capability to form films of sodium or calcium alginate and fibers of calcium alginates. Low-molecular weight alginates and oligo-alginates have better effects in various biological activities (Li et al., 2003). In particular, approximately 6-7 kDa alginate has antimicrobial effects and inhibitory effects on angiotensin-converting enzyme (Minoru et al., 2005). Treatment with oligo-alginate reportedly results in amelioration of cardiovascular disease and hyperlipidemia (Mao et al., 2004). Furthermore, oligo-alginate has been reported to inhibit the growth and differentiation of adipocytes, and the absorption of saturated fatty acids (Choi et al., 1986; Kim et al., 2010). Also, alginate is effective in the prevention of chronic diseases (Guven et al., 1991; Kim et al., 2010). Thus, the preparation of oligo-alginate from high-molecular-weight alginate is of interest (Lee et al., 2001). The traditional method for the preparation of oligo-alginate was primarily hydrolyzing alginates under acidic conditions with organic or inorganic acids, such as hydrochloric acid (Haug et al., 1967; Joo et al., 2003), sulfuric acid (Larsen et al., 2003), formic acid (Sherbrock-Cox et al., 1984), and oxalic acid (Haug et al., 1966). Although the procedure is convenient, common disadvantages of acidic hydrolysis include low recovery and partial saccharification in producing oligo-alginate (Kim et al., 2010). Moreover, the acidic hydrolysis of alginate is expensive because of the requirement or acid resistant reaction vessels and the neutralization of the acid used in the hydrolysis of alginates (Joo et al., 2003). To overcome the disadvantages of acid hydrolysis processing of alginate, alternative methods, such as radiation treatment, have been used in the hydrolysis of alginate (Joo et al., 2003). However, the most attractive method has been suggested to be enzymatic degradation, considering economic and environmental costs (Joo et al., 2003).
Alginate is degraded by alginate lyases through a β-elimination of the β-1,4-glycosidic bond. Two types of alginate lyases exist, which are specific for PG [poly-(1→4)-α-L-guluronate) lyase, EC 4.2.2.11] and for PM [poly-(1→4)- β-D-mannuronate) lyase, EC 4.2.2.3], and lyases also make 4-deoxy-erythro-hex-4-enopyranuronosyl groups at the nonreducing ends (Kim et al., 2010). Many studies have been performed using alginate lyases from microbes and lower animals to address the disadvantages of the traditional hydrolytic methods (Kim et al., 2010). However, to our knowledge, because of poor activity, stability, and specificity, no previous report has described an enzymatic reaction using alginate lyase. The objective of the current study was to isolate an alginate- degrading bacterium capable of degrading high substrate contents for commercial application in the future.
Sodium alginate was purchased from Sigma-Aldrich Co. (St. Louis, MO, USA) and other reagents were obtained from commercial sources.
>
Isolation of alginate-degrading bacteria
To screen alginate-degrading bacteria, seawater was collected off the northeastern coast of Korea. The seawater was diluted to 10, 100, and 1,000 times and spread onto PPES-II medium containing 1% alginate and 1.5% agar (Taga, 1968). Bacterial colonies grown on the plate after 7 days incubation at 25℃ were picked up and isolated for further study. For liquid culture, the strain was cultured in PPES-II medium containing alginate.
>
Identification of alginate-degrading bacteria
Bacterial strains isolated were identified by morphological, biochemical, and genetic characteristics as reported previously (Yu et al., 2012). Brief, a light microscope and a scanning electron microscope (SEM; model S-2400; Hitachi Ltd., Tokyo, Japan) were used for the morphological analysis. The VITEK Identification Kit (Biomerieux Inc., St. Louis, MO, USA) was used for physicochemical analysis. To perform genetical analysis, the 16S rDNA was amplified and sequenced. Then, homology search of sequences were conducted using a ribosomal database (http://www.ncbi.nim.nih.gov/BLAST/).
>
Bacterial growth and determination of alginate lyase activity
To investigate effects of temperature, pH, NaCl and alginate content on the growth of alginate-degrading bacterium, cells were cultivated in ranges of 10-50℃, pH 4-10, 0-10% NaCl, and 1-5% alginate, respectively. The abilities of alginate lyase from a single isolated bacterium were determined by analyzing the content of reducing sugar in the culture broth at the indicated incubation time. Decomposition of alginate polymers was checked at 254 nm using the Somogyi-Nelson method (Nelson, 1944; Somogyi, 1952).
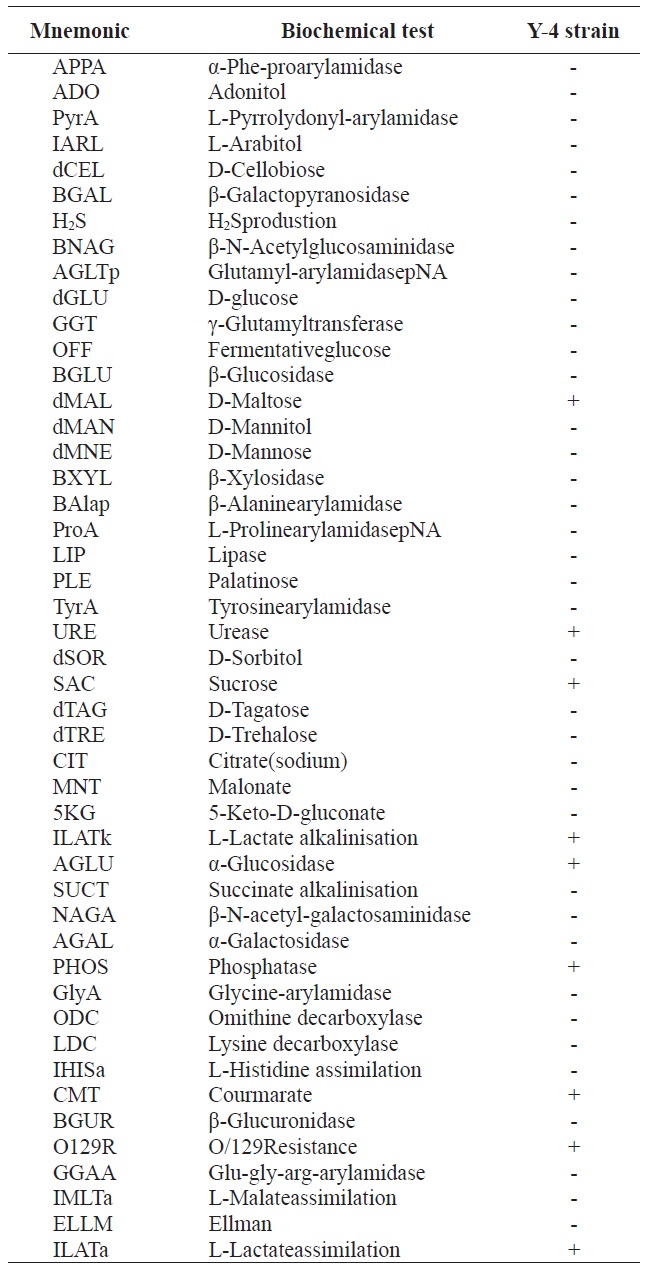
[Table 1.] Biochemical characteristics of alginate-degrading bacterium, Y-4 strain
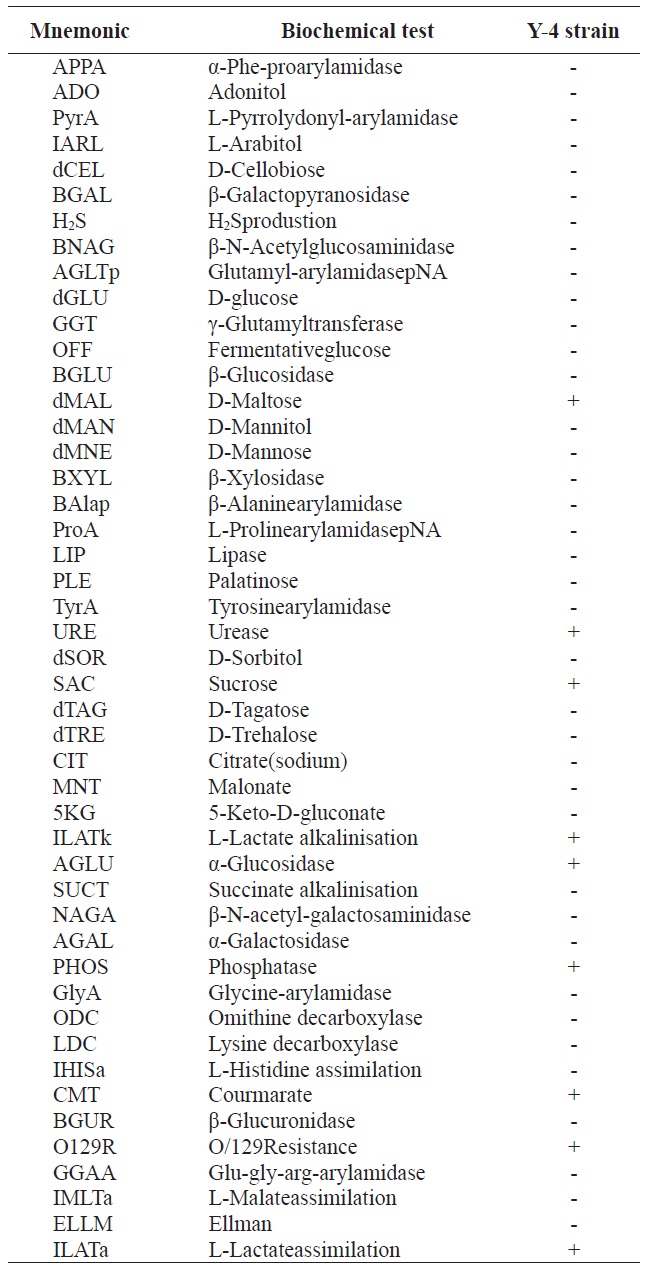
Biochemical characteristics of alginate-degrading bacterium, Y-4 strain
>
Isolation and identification of alginate-degrading bacteria from a marine environment
To obtain an alginate-degrading bacterium that used alginate as a carbon source, samples were spread and cultivated on agar plates as described in the Materials and Methods. After 7 days of incubation at 25℃, various colonies on the surface of PPES-II plate containing 1% alginate were chosen and colonies that formed a hole on the plate were taken for further study. Among them, a strain (
Gram staining and morphological analyses revealed that the Y-4 strain was Gram-negative and rod-shaped (data not shown). Biochemical characteristics of the Y-4 strain are listed in Table 1. However, these results provided limited information

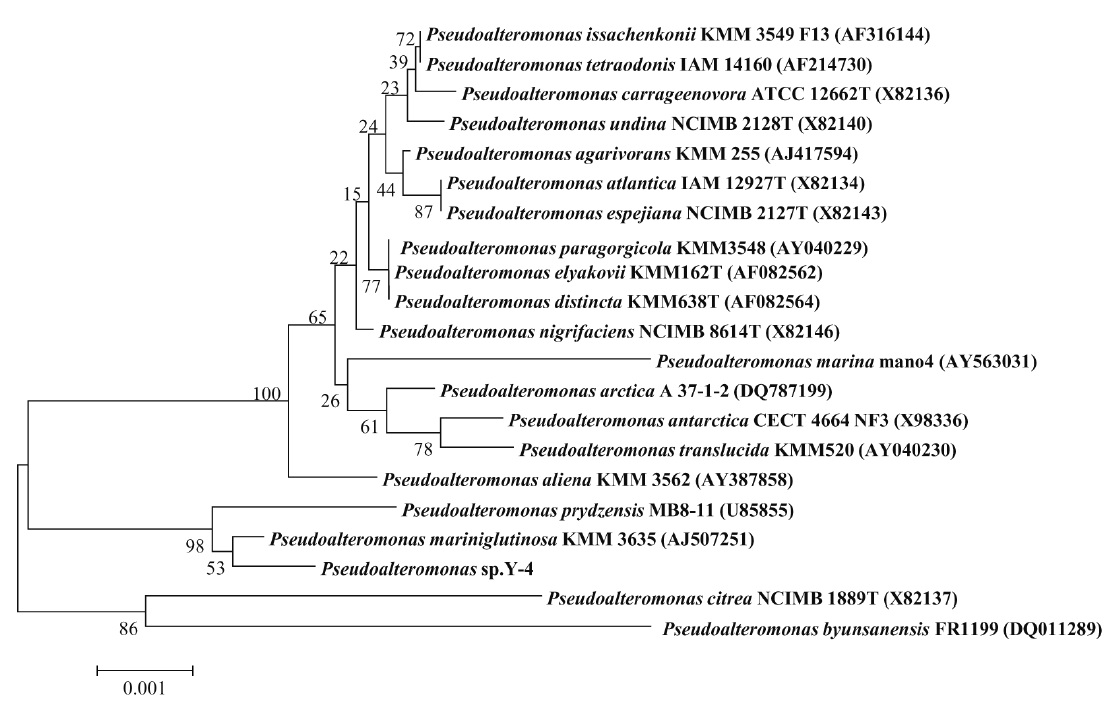
Identification of alginate-degrading bacterium, Y-4 strain, based on the homology search of 16S rDNA sequence
regarding the identity of the bacterium. Thus, we next conducted a genetic analysis using the bacterial 16S rDNA. A PCR product of 16S rDNA about 1.4-kb was obtained (data not shown) and the sequence was aligned with 16S rDNA sequences in GenBank using BLAST. As shown in Table 2, the isolated strain Y-4 exhibited 99% identity with another
>
Optimum culture conditions for alginate degradation by alginate-degrading bacterium
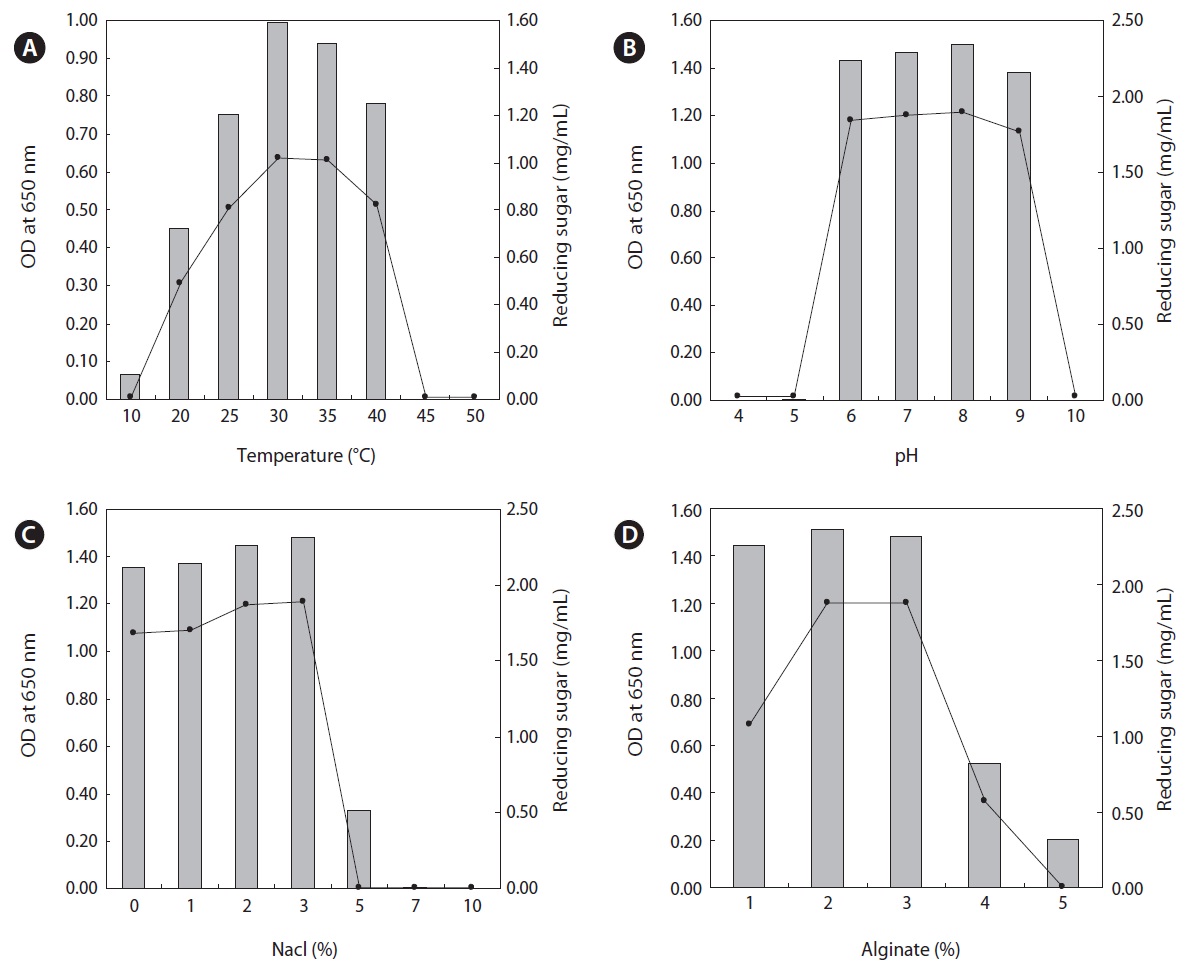
We investigated the effects of temperature, pH, NaCl, and substrate on the growth and alginate lyase activity of the Y-4 strain. The alginate lyase activity was evaluated by determining the content of reducing sugar in the culture broth as described in the Materials and Methods. The content of reducing sugars that originate from enzymatic degradation by Y-4 cells indicates the alginate lyase activity. The Y-4 strain was cultivated under conditions of pH 8.0, 2% sodium alginate and 2% NaCl with varying incubation temperatures. As shown in Fig. 2,
the Y-4 strain was able to grow in the range of 10-40℃ and the highest growth rate was observed at 30℃. The cell growth rate decreased below 20℃ and no cell growth was observed above 45℃. The highest content of reducing sugar resulting from the enzymatic degradation of alginate by the Y-4 strain was observed at 30℃ after a 24 h incubation (Fig. 2A). These results are consistent with other reports stating that marine bacteria capable of degrading alginate exhibited optimum alginate lyase activity in the range of 25-30℃ (Kim et al., 2010; Li et al., 2011b). Next, we investigated the effects of pH on bacterial growth and alginate lyase activity. The highest cell growth was observed at pH 8.0 and no significant difference was noted between pH 6.0 and 9.0 (Fig. 2B). No cell growth occurred below pH 5.0 or above pH 10.0. The highest content of reducing sugar was observed at pH 8.0 after a 24 h incubation (Fig. 2B). These results were similar to those reported by Kim et al. (2010). The highest cell growth and enzymatic activity occurred under conditions of 3% NaCl (Fig. 2C). Gradual retardation of cell growth was noted above 5% NaCl and no cell growth was observed above 7% NaCl, suggesting that the Y-4 strain was halophilic.
The most interesting finding was the effect of alginate concentration on cell growth and enzymatic activity. As shown in Fig. 2, the bacterium was able to grow in the range of 1-4% sodium alginate under conditions of 2% NaCl and pH 8.0 at 30℃. The optimum growth and the highest enzyme activity were observed in the presence of 2% and 2-3% sodium alginate, respectively (Fig. 2D). However, gradual retardation of cell growth and the activity observed over 4% sodium alginate and no cell growth occurred above 5% sodium alginate (Fig. 2D). Thus, the optimum culture conditions for the production of alginate-degrading enzyme were 2% sodium alginate, 3% NaCl, pH 8.0 and 30℃. Compared with previous results, the most notable characteristic of the Y-4 strains is its use of high substrate contents because most studies on alginate-degrading bacteria have reported that the optimum substrate concentration for cell growth was in the range of 0.5-1.0% alginate (Kim et al., 2010; Li et al., 2011a, 2011b). To our knowledge, this is the first report of the isolation of a marine bacterium degrading high concentrations of alginate (up to 4%). This result also suggests that the Y-4 strain possesses a notable gene(s) related to alginate degradation. To address this, further studies including cloning and expressing the genes are needed.