Ex situ conservation plays the most important role in the conservation of plants, especially conservation through botanical gardens or GenBank (Li et al. 2002). If wild populations become extinct, the ex situ-conserved populations maintain the evolutionary history and genetic diversity of the endangered species until the plant can be re-introduced back into natural habitats. Ex situ conservation is designed to maintain genetic diversity, to enhance the current population size, and to re-establish populations through restoration programs (Menges 1991, Schwartz et al. 2006). Estimation of the minimum size of the ex situ core collection that can capture the genetic diversity of the wild populations of endangered species is an essential component of efficient conservation strategies for reducing the risk of extinction of wild populations and for cost-effective genetic conservation programs (Brown and Briggs 1991, Palsbøll et al. 2006).
Brasenia schreberi J.F. Gmelin (water-shield) is a floating- leaved aquatic plant, which is found in marsh ponds, irrigation ditches, and lakes in the temperate and tropical regions of Asia, Australia, Africa, India, and North and South America (Cook 1996). In addition to having important functions in allopathic weed control (Elakovich and Wooten 1987), water-shield is also cultivated as edible plant in East Asia, where the young leaves are used in soups (Jiang and Cao 2008). Despite its ecological and economic utilities, the natural populations of the species are considered to be endangered in East Asia as a consequence of human activity and habitat loss (Wang and Chen 1994, Kim 1996). Previous studies have investigated the population genetics of B. schreberi in East Asia (Kim et al. 2008, Zhang and Gao 2008). Using randomly amplified polymorphic DNA (RAPD) and amplified fragment length polymorphism (AFLP) markers, the high level of genetic variation among the populations of B. schreberi in South Korea were related to their population sizes (Kim et al. 2008). However, the effective core collection size for the ex situ conservation of the water-shield plant is unknown, due to its limited sampling from natural populations. Water-shield plants combine sexual reproduction with vegetative propagation via rhizomes and winter buds (Chrysler 1938, Osborn and Schneider 1988). However, the rate of seed germination in the plant is too low to utilize dominant seeds for efficient ex situ conservation (Oh et al. 2008). As a perennial aquatic plant, the water-shield provides a model for the study of effective sampling for a species that primarily reproduces asexually.
Using AFLP markers, the aim of this study was to present the collection strategies that most efficiently capture the genetic diversity of B. schreberi. The AFLPs were selected because the markers produce a large number of polymorphisms rapidly, with few primer combinations and because they are highly reproducible and have good resolution (e.g., Archak et al. 2003, Kim et al. 2008, Na et al. 2010). This study provides genetic information for understanding B. schreberi, and thereby offers a genetic basis for ex situ conservation strategies for this kind of endangered species.
Our previous study on the genetic diversity of B. schreberi had indicated that among the six populations of South Korea, two populations (MGC from the mainland and JNS from Jeju Island) contained a relatively high degree of genetic variation (Kim et al. 2008). The MGC is the largest population from the mainland and is located on Cheonjin Lake of Goseong-gun, Gangwon-do. The JNS is a relatively small population at Galme Pond of Namjeju- gun, Jeju Island. A total of 140 ramets from these two populations (80 ramets from MGC and 60 from JNS) of B. schreberi in South Korea were examined in this study, using a simulation approach to determine the collection strategies that most efficiently capture the genetic diversity of the plant for ex situ conservation. To avoid sampling the same plants, the distance between individual specimens collected within a given population was at least 5 m. All of the leaf tissues were stored on ice until shipment to Ajou University. Upon arrival, the samples were rinsed in deionized water, spilt into 200 mg aliquots, and were stored at -70℃ until DNA extraction.
Total genomic DNA was isolated from leaf tissues (0.2 g) following the procedure for rapid DNA minipreparation (Chen and Ronald 1999). The extracted DNA was resuspended in Tris-EDTA buffer (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA [pH 8.0]), and the DNA concentration was determined spectrophotometrically (Gene flow Ltd., Staffordshire, UK).
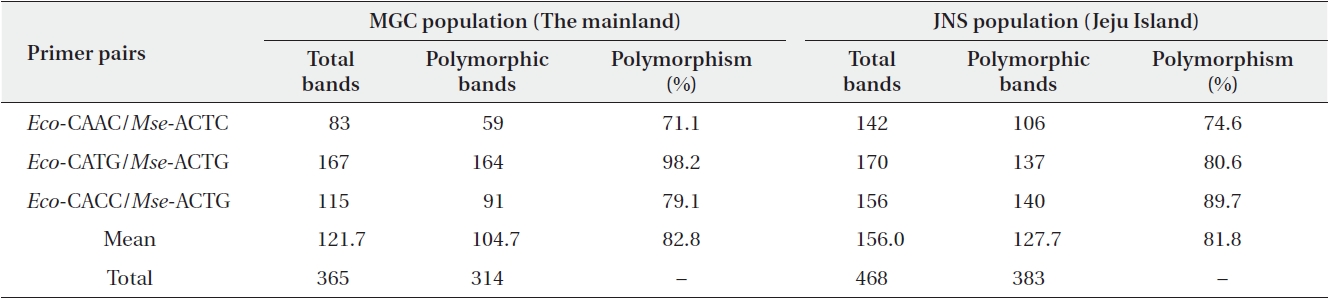
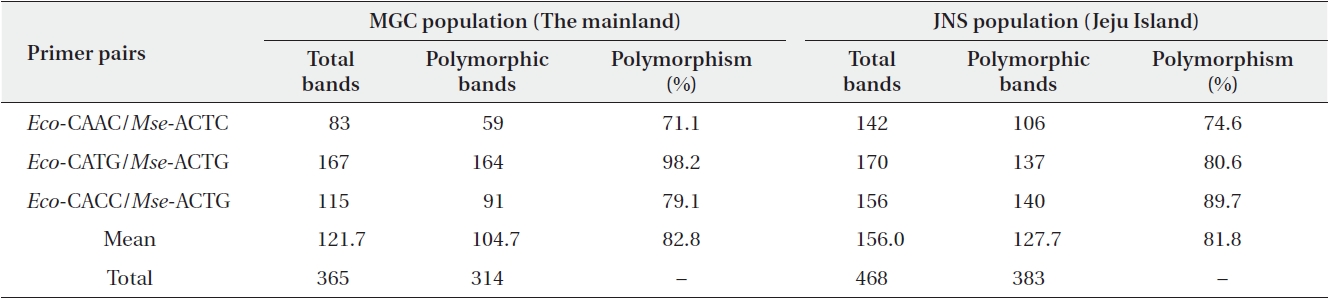
AFLP analysis was performed following the protocol established by Vos et al. (1995), and the detailed procedures are outlined in Kim et al. (2008). Briefly, the analysis was carried out using Eco-C and Mse-A as pre-selective primers. The pre-amplification products were diluted 10-fold with distilled water and were used as the template for further selective amplification using selective primer combinations, Eco-CAAC/Mse-ACTC, Eco-CATG/Mse-ACTG, and Eco-CACC/Mse-ACTG; the selective amplification was performed with Eco primers tagged with a 6-FAM dye (Table 1). The AFLP alleles were resolved using an ABI 3100 genotyper (Applied Biosystems, Foster City, CA, USA) with GENESCAN 3.7 (Applied Biosystems) and LIZ600 as an internal lane size standard.
The AFLP bands were scored as ‘1’ (present) or ‘0’ (absent) in a binary matrix for each primer, and monomorphic bands across all individuals were discarded from further analysis (Keiper and McConchie 2000). For esti-
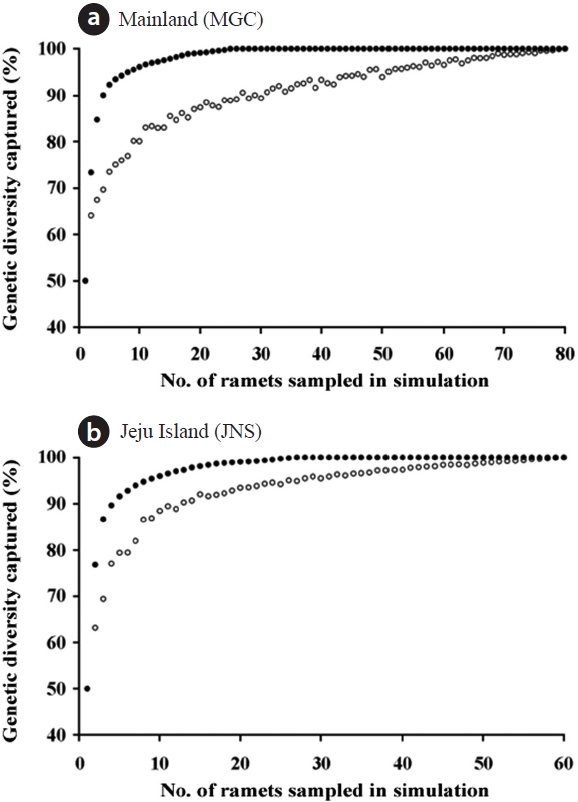
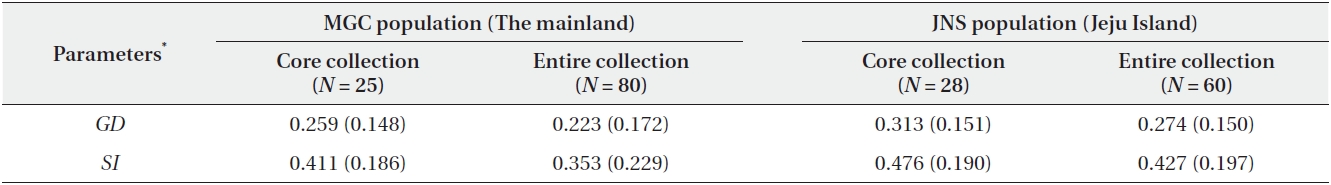
mating the minimum population size of B. schreberi, we employed a sampling method using the maximization strategy (Schoen and Brown 1993) that generated core collections to maximize the level of genetic diversity captured. The simulations of sampling by the maximization strategy were performed using MSTRAT 4.1 (Gouesnard et al. 2001). The efficiency of the sampling strategy was assessed by comparison with the level of genetic diversity captured in the simulation of simple random strategy without replacement using MSTRAT. The sample size in each step of the simulations was increased by one ramet, and 20 independent replicates were simulated in each step. The minimum population size for the ex situ conservation of B. schreberi was determined as the minimum number of ramets necessary to capture all of the polymorphic bands of AFLP markers (i.e., the number of ramets in the core collection). We calculated Nei’s (1973) genetic diversity (GD) and Shannon and Weaver’s (1949)
index (SI) for the core and entire collections using POPGENE 1.31 (Yeh et al. 1997).
Using the three primer combinations, a total of 365 bands were generated for the MGC population. Of these, 314 (86.0%) were polymorphic bands, which ranged from 59 to 164 bands with an average of 82.8 bands. In Jeju Island population (JNS), these primer combinations resulted in 468 amplification products. Of these, 383 (81.8%) were polymorphic (Table 1). All of the 140 individuals sampled were distinguished by distinct AFLP phenotypes. The data matrix of allelic frequencies showed that a large proportion of rare alleles (with a frequency < 0.05) were detected in the MGC population (18.2%) and JNS population (14.2%) (Fig. 1).
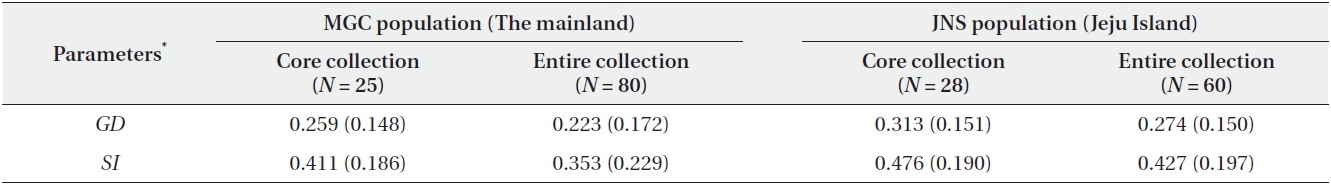
The simulation of sampling by the maximization strategy showed that a sample of 25 individuals (31.3%) captured all of the polymorphisms in the mainland population. In the case of the Jeju Island population, a sample of 28 individuals (46.7%) included all of the maximized genetic diversity (Fig. 2). The core collection of this study included all of the AFLP alleles of the entire collection. Furthermore, the values of GD and SI were similar between the core collections and the entire collection (Table 2).
The sampling size and selection of populations for the ex situ conservation of endangered species is primarily aimed at the preservation of the species, principally from threats due to human activity (Brown and Briggs 1991, Li et al. 2002, Hoban et al. 2010). In the maintenance program of an endangered species, the use of molecular data as the primary approach to defining the conservation size is appropriate, as this approach provides information on the current distribution of genetic diversity and the history of the population (Milligan et al. 1994, Schwartz et al. 2006, Richards et al. 2007). Using combined nuclear ribosomal internal transcribed spacer and chloroplast trnL-F sequence data, we have found that the populations of B. schreberi were subdivided into two groups in South Korea: those of the mainland and of Jeju Island. A low level of population differentiation was also detected within each region (Kim et al. 2012). Thus, for the present study, we used AFLP makers to determine the population size for ex situ conservation in the two populations (the MGC population [mainland of South Korea] and the JNS population [Jeju Island]). These two populations were found to contain a relatively high genetic diversity and a large population size, and the polymorphisms of the AFLP marker were more frequently detected than those produced by RAPD in a previous study (Kim et al. 2008).
Based on the maximization strategy for estimating the minimum population size, we determined that 28 individuals (46.7%) contained the total genetic diversity captured in the JNS population, whereas 25 individuals (31.3%) contained the total genetic diversity captured in the MGC population (Fig. 2). The maximum strategy used in this study captured 100% of the allelic diversity existing in the entire collection. Moreover, the levels of genetic diversity of the core sets were similar to those of entire collection (Table 2). These data indicated that the core sets were a very good representation of the entire collection. The present core sets, maximizing the genetic diversity, also reduced the collection size for both populations. The efficiency of the maximization strategy was well shown by the study of endangered plant Aster altaicus var. uchiyamae (Nakai) Kitam. (Kim et al. 2011). In their study, the minimum population sizes of three populations were reduced to 13%, 33%, and 36%, respectively, compared to the random sampling from A. altaicus var. uchiyamae (Kim et al. 2011).
When compared to what has been described for other aquatic plants, our data showed that a relatively large number of individuals would be required for the ex situ conservation of B. schreberi in South Korea. For example, Richards et al. (2007) had calculated that only 13% of the Zizania texana population, an aquatic plant with asexual propagation as the primarily reproductive mode, could comprise all 70 of the alleles, as based on six microsatellite markers. This result can be attributed to the reproductive mechanism of Z. texana. Although the vegetative propagation occurring via rhizomes of B. schreberi is similar to that of Z. texana, B. schreberi can also propagate via winter buds, which can have a relatively extensive dispersal (Liu et al. 2005). The difference in polymorphisms between AFLP and microsatellite markers may also have increased the number of individuals required for the ex situ conservation in B. schreberi. All of the 140 sampled ramets were distinguished as distinct genets by AFLP markers.
Eventually, the goal is that captive populations can be released back into natural or restored habitats. The main objective of ex situ conservation is to maintain the species in captivity until habit restoration allows its release back into nature. However, during the process of ex situ conservation, various degrees of reduction of genetic diversity have been reported in captive populations relative to their natural populations (Wright and Gaut 2005). Therefore, further work should investigate the genetic diversity within captive and natural populations to evaluate the sampling strategy for the ex situ conservation of B. schreberi populations. The optimal breeding program of core collections should be designed to avoid the potential risk for reducing the genetic diversity of core collections.
In conclusion, our sampling model was designed to assess the maximum amount of polymorphism within the population. These initial surveys may be helpful to reduce the time and cost of collection efforts. Our population genetic data may be a key guideline for the determination of optimum sampling sizes in ex situ collection, which will ultimately depend on the genetic structure of B. schreberi populations. Although the size of the population chosen for ex situ conservation would depend on the genetic structure between regions, the optimal ex situ collection should include replicate samples from genetically similar populations within geographical distinction. The combination of our genetic data with sufficient knowledge of ecological and demographic information will provide a clear guide for maximizing the genetic diversity within any conservational collection.





![Histograms of the allelic frequencies for (a) 314 (mainland population [MGC]) and (b) 383 polymorphic bands (Jeju Island population [JNS]).](http://oak.go.kr/repository/journal/11834/STHHCL_2012_v35n4_301_f001.jpg)