The goal of image registration is to find out the geo-metric transformation of two or more images of the same scene that are taken at different instances, from different sensors or from different viewpoints and align them into the same spatial coordinate. The alignment is done by taking one image as
Many registration techniques have been developed for the various medical imaging problems. The development of effective registration techniques has been demanded for efficient clinical applications. Registration can be divided into rigid or non-rigid, according to the nature of the transformation [2]. Rigid registration is simpler, as its transformation in three dimensions is defined by six parameters: three for rotations and three for translations. Conversely, non-rigid registration is more complex. Besides, rotations and translations it involves voxel-dependent dis-tortion. Although rigid transformation is simple compared to non-rigid transformation, it is a crucial task for regis-tering rigid structures such as bones, and it is often per-formed as a preliminary step for some non-rigid registration [3, 4].
From the aspect of the type of features that are used in a registration, the existing automatic registration methods are further divided as feature-based or intensity-based methods. Feature-based methods extract the corresponding features or attributes between a reference and the float images for registration. Common features include points, edges, contours or surfaces. These features may be specified manually or extracted automatically. Among the feature-based rigid registration methods, the principal axis regis-tration (PAR) method [5] is the most motivating one as it can easily find the transformation parameters by calculat-ing the principal axis of a common feature that is extracted from a reference and the float images.
PAR registration consists of translating the centroid of a float onto the reference image, and then, rotating it about the centroid to align the 3D principal axes of both the images. Hence, good features need to be extracted to obtain the accurate principal axes and centroids. In most of the 3D cases, a binary structure image [6, 7] is used as features while the contour features [8] are extracted for 2D principal axis registration. The main advantages of the feature-based principal axis registration methods are simple, and they are less computationally complex. Despite good properties, these approaches have been less popular as they can lead to inaccuracies in a medical image registration.
Intensity-based methods that uses full image content is the most interesting work, as they provide more reliable results with higher accuracy and this makes thees meth-ods suitable for clinical application [9]. These approaches are commonly known as the voxel similarity based meth-ods for 3D cases. These methods solve the registration task as a mathematical optimization problem. They use a similarity measure as a cost function to gauge the quality of the alignment between the reference and the float images for any given transformation. Obviously, finding out the optimized parameters between the two images needs the maximization of the similarity or the minimiza-tion of the difference. Common similarity metrics are mutual information (MI), normalized mutual information (NMI), cross correlation (CC), normalized cross correlation (NCC), sum of squared differences (SSD) and sum of absolute differences (SAD) [10, 11]. MI or NMI is consid-ered as the most suitable metric for the multimodal image registration [12, 13] while the rest of them are appropri-ate for mono-modality image registration [14-16].
There are many mathematical optimizers that are sug-gested for image registration. Common optimizers include Powell’s, downhill simplex method, steepest descent, quasi-Newton, Levenberg-Marquardt, and conjugate-gra-dient method [17]. These approaches usually apply the voxel value of original intensity images and use an opti-mizer. Gradient NMIs based registration has been proposed for the registration of 3D CT data to 2D endoscopic image [18]. This approach calculates the NMIs in the gra-dient images as well as in the original intensity images and it adopted two optimizers, Powell’s and downhill simplex method. The voxel similarity method has been known to provide high accuracy for clinical application but due to their high computational cost, their usage in 3D clinical applications has been limited.
In order to diminish the computational complexity of the voxel similarity based methods, multi-resolution approach has been developed by using the hierarchical structure of the input images [19-21]. The hierarchical structure is constructed by consecutively subsampling the pyramid of two input images. The registration is fulfilled from low to high resolutions in the pyramid. The transformation parameters are searched in the optimization process, starting from the maximum similarity score or minimum difference between the two volumes at the lowest resolu-tion. Then, the parameters obtained from the first (or low-est) resolution level are passed to the next resolution level. The registration is performed until the highest reso-lution level of the pyramid is reached, where the optimal parameter is achieved. As the level increases, the sub-sampling interval decreases, and the accurate registration result is obtained with a higher computational complex-ity. Many toolkit and toolboxes have been progressively developed for the voxel similarity method and for the multi-resolution methods [22, 23]. Hence, the computa-tional complexity of the multi-resolution approach is reduced compared to the original voxel similarity method by using full image content. However, the accuracy of the registration depends on the definition of the level of the pyramid structure. The computational cost for a 3D vol-ume is still an issue for real-world medical image appli-cations.
Equivalent meridian plane (EMP) based MI registra-tion method [24, 25] integrates the feature-based and voxel based methods to register the multi-modal brain volumes, where the volumes are roughly aligned on the basis of the EMP which is determined by principal com-ponent analysis (PCA). Then, the registration is refined by maximizing the MI between the EMP of a float vol-ume and the 2D XY-coordinate plane of the reference volume. Powell’s optimization method is performed for the enhancement of searching during the registration pro-cess. Even though this method is fast thanks to the 2D-plane based registration, the registration result depends on the correctness of EMP. This infers that it cannot guar-antee an optimal solution for every case.
The purpose of this study is to provide a novel method for a rigid registration of 3D CT images. The registration domains in this study are rigid, three-dimensional, and mono-modal images of the same patient. The proposed method aims to combine the advantages of the feature-based and the voxel similarity based methods for the enhancement of the overall performance in the aspects of both accuracy and computational cost. Section II describes the methodology of the proposed method in detail. Experimental results and analysis are presented in section III. Finally, conclusions are drawn in section IV.
II. PROPOSED FEATURE-VOXEL BASED IMAGE REGISTRATION
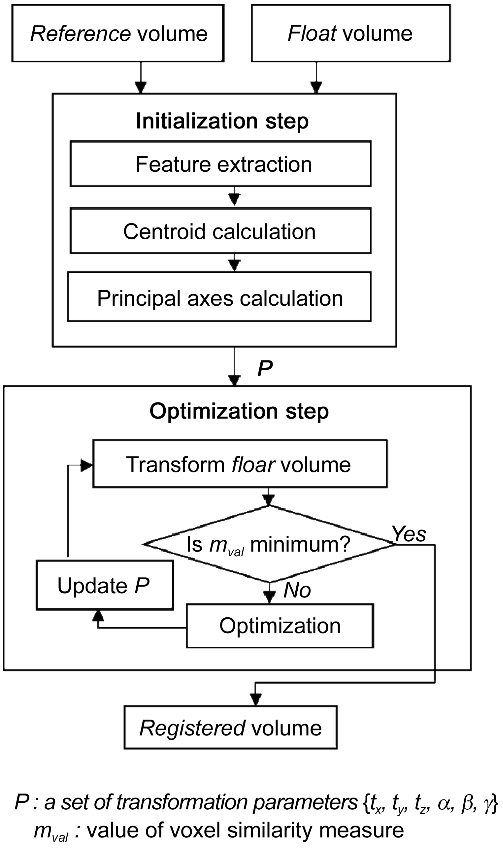
The proposed feature-voxel based image registration method consists of two steps: initialization and optimiza-tion step as shown in Fig. 1. In the initialization step, six transformation parameters are calculated based on the centroid and the principal axes of two CT volumes. The
estimated parameters are passed to the optimization step in order to obtain the optimal parameters. Differing from the previous EMP-MI approach, the proposed method uses the whole 3D volume in order to provide trustworthy registration result. It calculates the NCC score between the reference and the float volume by using the downhill simplex method.
>
A. Estimation of Initial Transformation Parameters
Let
Where,
Hence, the vector representation of a rigid body trans-form of Equation (1) can be rewritten as,
A common way to describe a rotation matrix is the decomposed form of a sequence of the following matrices
Where,
the reference volume
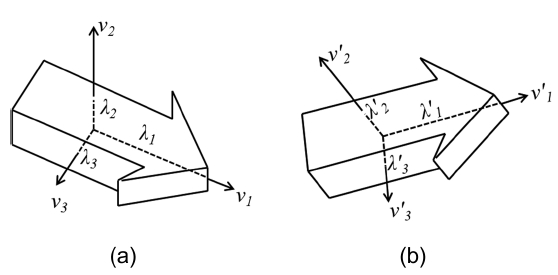
The initial parameters of the two volumes are com-puted based on the transformation of their principal axes. Fig. 2a illustrates a 3D object with its principal axes where eigenvectors,
A binary volume is used as the feature to represent the geometric shape of 3D to extract the principal axes. Let
Where,
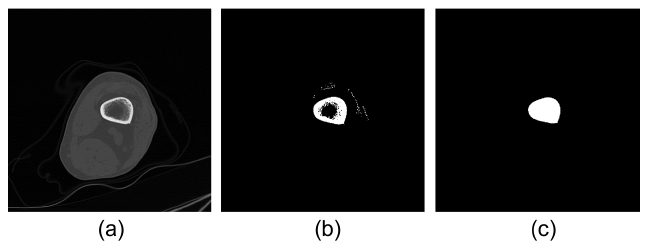
The bone is the most special feature in CT images. Therefore for knee CT registration, the bone region must be extracted from unwanted tissues, such as muscle or fat. The global threshold value, τ, of the bone mask is chosen as 300 Hounsfield units (HU) for the intensity value range of CT image from -1,024 HU to 2,163 HU. Fig. 3b shows the candidate bone region with some resid-uals that are to be removed and holes that are to be filled after thresholding. The residuals are removed by 3D labeling and the holes are filled by the morphological operation. Fig. 3c shows the feature of a bone region that will be used for registration.
The centroid of
The principal axes of the binary object are defined by the eigenvectors of its inertia matrix [6]. In fact, inertia matrix is the matrix that can be composed by the vari-ances and covariance between the axes. The inertia matrix,
Where,
and
Here,
After the three eigenvectors are calculated and form the matrix
To calculate a set of six parameters
Step 1: Define the binary volumes
Step 2: Compute the centroid
Step 3: Calculate inertia matrixes
Step 4: Extract the rotation angles
Step 5: Take the difference of
Step 6: Compute the subtraction between
After the computation of the initial transformation param-eters, which approximately estimates the coarse result to the ideal value, the task that remains is to find out the optimal transformation parameters for the two volumes. The process of finding out the optimal parameters that involves the computation of the voxel similarity mea-sures between the reference and the float volumes. It is followed by the optimization step. The purpose of the similarity measure is to return a value that indicating how matched the two images are [9]. The cost function to determine the optimal parameter matrix is defined by,
Where,
Where,
represent the mean intensity values and σr and σf are the standard deviation of the respective vol-umes. They can be computed as,
In principle, the final transformation parameters are considered optimal when the NCC score between
>
A. Experimental Setup and Evaluation Method
The efficacy of the proposed method is evaluated on both phantom and synthetic data sets. As the purpose of this study is 3D rigid medical image registration, the phantom was simply created in a rigid structure. Fig. 4 describes the phantom that consisting of three erasers inside a box and mounted on a rectangular acrylic plate. The phantom was scanned in a CT scanner provided by Ajou University Hospital, Korea. The reference volume was first scanned without any transformation of the plate. Five floating volumes were obtained by scanning with a random transform of the phantom in x-, y-, z- axes. It is illustrated in Fig. 5. Each CT volume is Digital Imaging
and Communications in Medicine (DICOM) image type. It had a dimension of 512 × 512 × 279 and slice thickness of 1 mm, with a 16-bit depth and 0.6836 mm pixel spac-ing. One particular medical CT volume of knee acquisi-tions was selected for the synthetic data study. We created a database of transformed volume by applying a rigid transformation to it. The knee CT volume had a 16-bit depth, 512 × 512 × 48 in dimensions, 3 mm in slice thick-ness and 0.6836 × 0.6836 mm in pixel spacing. The mag-nitude of the rigid transformations varied between ±10 degree for rotation and ±10 pixel for translation. Within these ranges, 40 volumes were randomly generated to validate the registration.
The proposed method is compared with a voxel simi-larity measure based approach (voxel-based), and a multi-resolution based approach. The voxel-based method establishes a voxel-to-voxel mapping between the refer-ence and the float volume. On the other hand, the multi-resolution registration reduces the computational com-plexity of the voxel-based registration method which is accurate but at the same time it is time-consuming. The 4-level volume pyramid for multi-resolution registration is obtained by resampling both the reference and the float volume along each
called from the MATLAB (MathWorks, Natick, MA, USA) as mex file. The remaining parts of the programs were implemented in MATLAB 7.11 and they run on a PC with Intel® Core™ i7, 2.67 GHz processor and 8 GB RAM. The registration time, excluding the time of the loading data sets, is in seconds.
NCC scores between the reference and the registered volumes are computed for the evaluation of the accuracy of the proposed method on phantom images. The closer the score is to one, the higher the accuracy of the registra-tion of the two volumes. In the case of synthetic image registration, the accuracy is measured by the mean and standard deviation in the differences between the com-puted transformation parameter values (
>
B. Registration on Phantom Data Sets
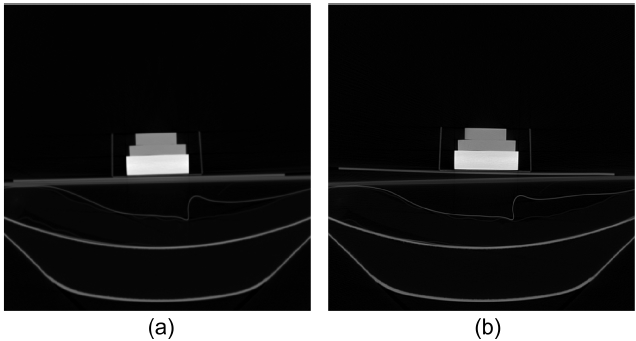
Registration of the phantom is performed while setting up the first scan as the reference volume and the remain-ing five scans as float volumes. The reference volume is named as set_1 and the remainders are serially named as set_2 to set_6. In order to accomplish registration, the appropriate volume of interest (VOI) is selected from both the reference and the float volumes. The size of VOI for each volume is 231 × 131 × 111 and the registration process is taken on those VOIs. Fig. 6a and 6b show slices of VOI of the original reference and float volumes. For the phantom study, the binary volume which is needed for the extraction of the principal axes is simply obtained by using Otsu [27] thresholding method that is followed by 3D labeling. Otsu’s method [27] is a non-parametric and unsupervised method that provides an automatic global threshold value τ in Equation (4). Fig. 6c and 6d shows the binary results after the application of the Otsu’s method [27]. Some unwanted residual remains apart from the desired region. Those residuals are removed by labeling the connected component of the 3D binary volume. Fig. 6e and 6f show the binary volumes that are ready to extract the principal axes, after the labeling pro-cess. Fig. 6g and 6h show slices of the segmented regions for reference and float volumes, respectively.
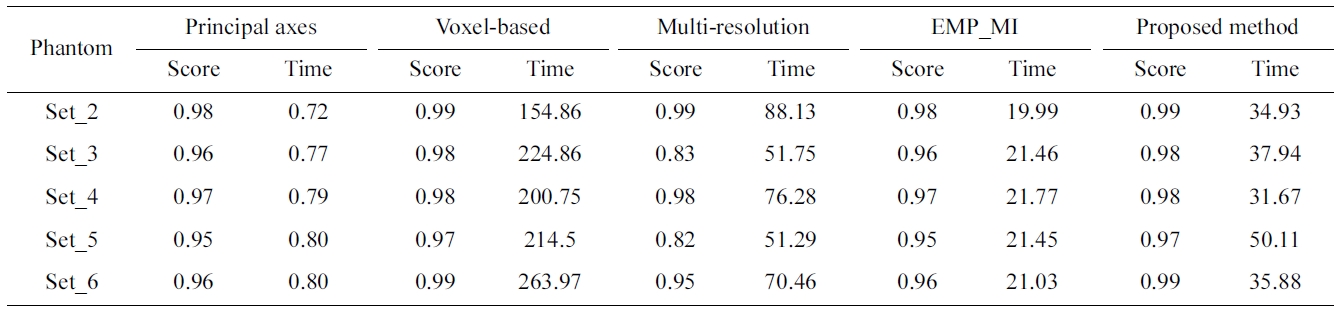
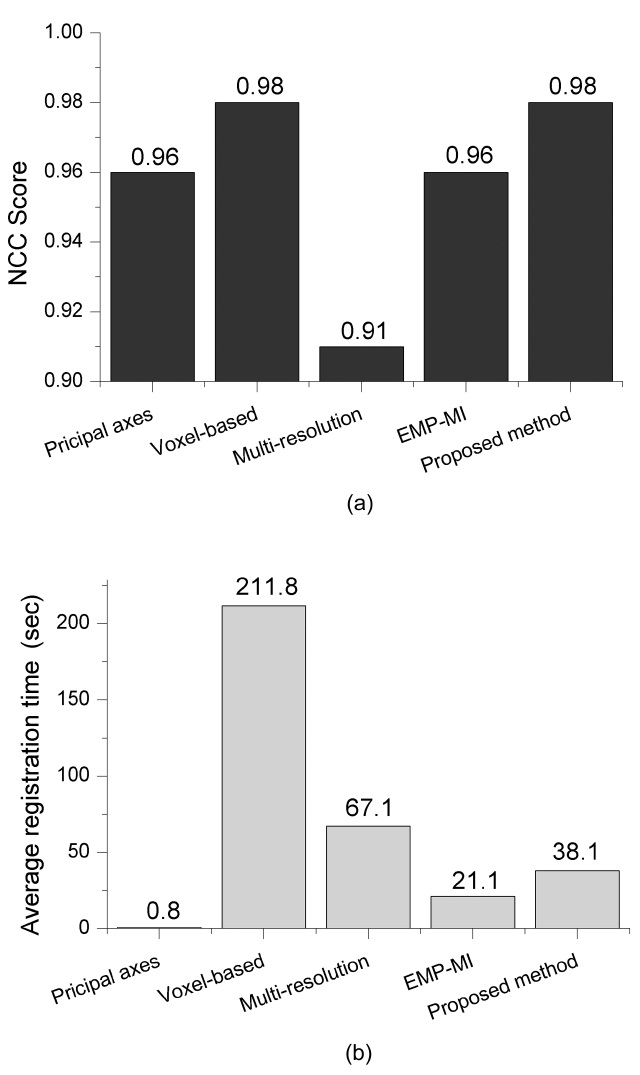
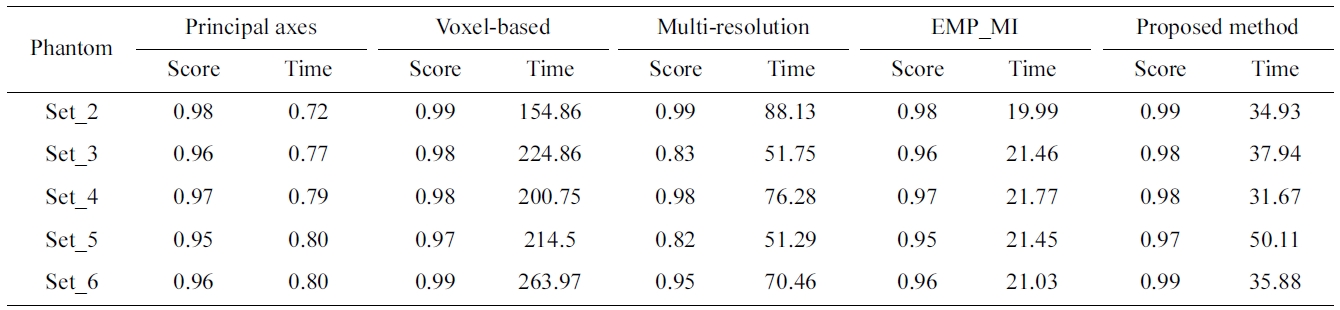
The superimposed 3D visualization of the registration results for the reference (set_1 in dark gray) and the float (set_6 in light gray) volumes are presented in Fig. 7. Table 1 compares the NCC score and the processing time of the proposed method against the principal axes based approach, the voxel-based, the multi-resolution based method and EMP based MI method.
The average NCC score and registration time are illus-trated in Fig. 8. The figure reveals that the voxel-based approach provides high accuracy result with high compu-tational complexity. Multi-resolution based approach reduces the computational cost but the average NCC is low. This implies that it may produce unsatisfied results.
Correlation score and registration time for phantom data sets (set_1 as reference and the remaining volumes as float) : time insecond
Both the principal axes based approach and the EMP-MI based approach provide a fast processing time result but a low correlation score is obtained. Although the proposed method is not as fast as the other two methods, it provides
[Table 2.] Evaluation of the proposed method with different measures
Evaluation of the proposed method with different measures
Mean (standard deviation) of the transformation error measurement of forty synthetic data sets
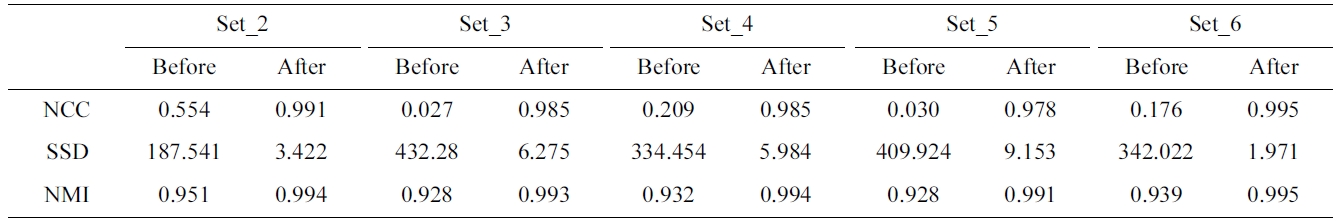
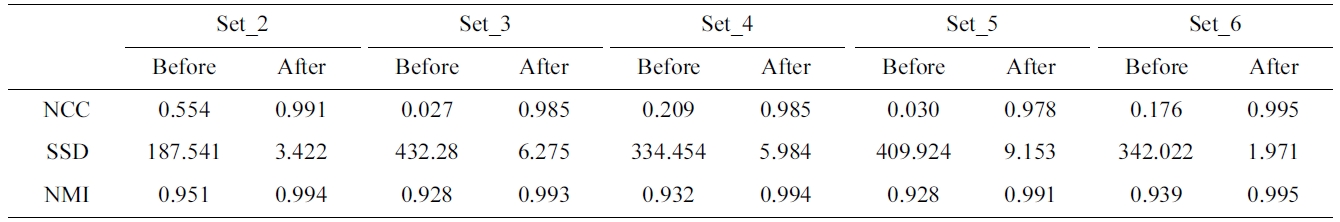
efficient registration result as the average NCC score is as high as the voxel-based method. Moreover, Table 2 describes the before- and after-registration score of dif-ferent similarity measures for each phantom data set.
As NCC, the higher the score of NMI, the higher is the accuracy of the registration. In contrast to NCC and NMI, the higher accuracy of registration is obtained when the score of SSD is small. To evaluate the proposed method with different similarities, the correlation score of the ref-erence (set_1) and the registered volume which is trans-formed the float volume (set_2-set_6) by the parameters resulted from different similarity measures. The average correlation score of NCC and NMI is 0.98 and the aver-age correlation of SSD is 0.96. The average registration time of NCC is 38.1 seconds, while those of NMI and SSD are 53.2 and 23.7 seconds, respectively. Therefore, NCC provides the similar average score with NMI, higher score of SSD and it acquires a faster average regis-tration time.
>
C. Registration on Synthetic Data Set
The registration is also evaluated on the synthetic datasets that are randomly generated by rotating and translating one knee CT volume along the
A set of new float images were created by the applica-tion of the random rigid transformation with a rotation that varies from -10 degree to +10 degree and a transla-tion which varies from -10 pixels to +10 pixels along all the axes. As the registrations are performed with the ground-truths, the accuracy is evaluated by the mean and
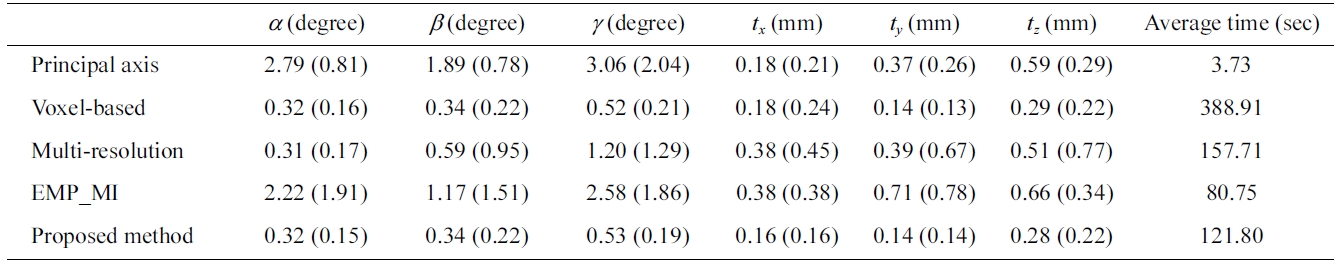
standard deviation (SD) of rotation and translation errors. Standard deviation shows the amount of variation from the mean. The smaller the standard deviation, the higher is the accuracy of registration. Table 3 shows the mean (SD) of the transformation errors and the average pro-cessing time in the registration of the set of new float images with the reference images by different approach. Although the processing time of the principal axes based approach and the EMP-MI based approach is faster than the voxel-based, multi-resolution based and the proposed methods, they provide a big mean (SD) value of synthetic data sets. The proposed method provides more accuracy result and relevant processing time of registration. Fig. 9 shows the registration result of one of the synthetic data sets.
IV. CONCLUSIONS AND FUTURE WORK
This paper presents a novel method to automate the process of registering 3D CT images taken at different instances. The proposed two-step method first finds out the initial transformation parameters based on the princi-pal axes of the binary structure of the input images. The parameters acquired in the first step are then optimized to provide the parameters that best align the float volume to the spatial coordinates of the reference volume. The objective of this preliminary study using phantom and synthetic data is to validate the performance of the pro-posed method in terms of accuracy and processing time. The experiments on phantom and synthetic data show that the algorithm provides robust and faster results com-pared to the voxel similarity measure based approach and the multi-resolution registration method. The experimen-tal results demonstrated that the proposed method can be applied for the rigid registration of real images. In future, real patient data sets are to be examined. Further, a more robust segmentation method is to be employed to obtain better results for the patient images, as the current corre-lation calculation is limited only to the bone region.

![Slice 40 of (a) original reference volume (set_1) (b) original float volume (set_6) (c) reference binary volume and (d) float binary volume after the Otsu’s method [27] (e) referencebinary volume and (f) float binary volume after 3D labeling (g) segmented reference volume (h) segmented float volume.](http://oak.go.kr/repository/journal/11652/E1EIKI_2012_v6n1_1_f006.jpg)