MEDLINE® citations are indexed using the Medical Subject Headings (MeSH) ® controlled vocabulary. This indexing is performed by a relatively small group of highly qualified indexing contractors and staff at the US National Library of Medicine (NLM). Their task is becoming more difficult, due to the yearly increase of MEDLINE, currently increasing by around 700 k articles per year [1].
The Medical Text Indexer (MTI) [2-4] is a support tool for assisting indexers as they add MeSH indexing to MEDLINE citations. MTI has two main components: MetaMap [5], and the PubMed® Related Citations (PRC) algorithm [6]. MetaMap analyzes citations, and annotates them with Unified Medical Language System (UMLS) ® concepts. Then, the mapping from UMLS to MeSH follows the Restrict-to-MeSH [7] approach, which is based primarily on the semantic relationships among UMLS concepts (MetaMap Indexing, MMI). The PRC algorithm is a modified k-nearest neighbor (k-NN) algorithm, which relies on document similarity to assign MeSH headings. The output of MMI and PRC are combined by linear combination of their indexing confidence. This method attempts to increase the recall of MTI, by proposing indexing candidates for MeSH headings that are not explicitly present in the title and abstract of the citation, but that are used in similar contexts. Finally, a post-processing step arranges the list of MeSH headings, and tailors the output to reflect NLM indexing policy.
We are studying the use of machine learning, to improve the MeSH heading assignment to MEDLINE citations performed by MTI. While comparing and selecting indexing methods is manageable with a limited number of MeSH headings, a large number of them make automation of this selection desirable.
In this work, we present a methodology to automatically select an indexing algorithm for each MeSH heading. Experiments are performed on the whole set of MeSH headings, and on a set of MeSH headings known as Check Tags [8]. Check Tags are a special class of MeSH headings routinely considered for every article, which cover species, sex and human age groups, historical periods, and pregnancy. We show that this process can be automated, based on previously indexed MEDLINE citations.
We find that most of the existing MeSH indexing methods fit into either pattern matching methods, which are based on a reference terminology (like UMLS or MeSH), or machine learning approaches, which learn a model from examples of previously indexed citations.
In the machine learning community, the task of MeSH indexing has been considered as a text categorization problem. Publication of the OHSUMED collection [9], containing all MEDLINE citations in 270 medical journals over a five-year period (1987-1991) including MeSH indexing, provided a large body of data that enabled us to view MeSH heading assignment as a classification problem. The scope of the collection determined the subset of MeSH that can be explored. For example, Lewis et al. [10] and Ruiz and Srinivasan [11] used 49 categories related to heart diseases with at least 75 training documents, and Yetisgen-Yildiz and Pratt [12] expanded the number of headings to 634 disease categories. Poulter et al. [13] provide an overview of these and other studies of classification methods, applied to MEDLINE and MeSH subsets.
Among the pattern matching methods, we find MetaMap, as mentioned above, and an information retrieval approach by Ruch [14]. Pattern matching considers only the inner structure of the terms, but not the terms with which they co-occur. This means that if a document is related to a MeSH heading, but the heading does not appear in the reference source, it will not be suggested.
Currently, MeSH contains 26,142 main headings and over 172,000 entry terms, to assist the indexers in determining the appropriate main headings to assign to a MEDLINE citation. Small-scale studies with machine learning approaches already exist [12, 15]. But the presence of a large number of categories has forced machine learning approaches to be combined with information retrieval methods designed to reduce the search space. For instance, PRC and a k-NN approach by Trieschnigg et al. [16] look for similar citations in MEDLINE, and predict MeSH headings by a voting mechanism on the top-scoring citations. Experience with MTI shows that k-NN methods produce high recall, but low precision indexing. Other machine learning algorithms have been evaluated that rely on a more complex representation of the citations, e.g., learning based on Inductive Logic Programming [17]. In previous work [18, 19], we showed that MeSH headings have different behavior, depending on the indexing algorithm used.
The selection of the best indexing method is a challenging task, due to the number of available categories and methods. In this paper, we present a methodology that automates the selection of indexing algorithms based on meta-learning.
In machine learning, meta-learning [20, 21] applies automatic learning to machine learning experiments. In our work, the experimental data are indexing algorithm results, which are used to select the most appropriate algorithm.
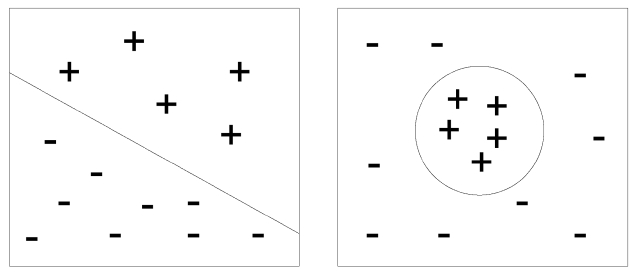
Indexing methods have different performance, depending on the MeSH heading. To illustrate why this happens, we can place the citations in a two dimensional space, in which a + sign is a positive example, and a - sign is a negative example.
Fig. 1 shows two sets of instances represented in this vector space. In the left image, the positive and negative citations can be split into two sets, based on a separating hyperplane, supporting the use of a support vector machine (SVM) approach with linear kernel. In the right image, it is not possible to identify a hyperplane, so another kind of learning algorithm is required, e.g., k-NN or SVM with non-linear kernel.
Without previous experimentation, it is difficult to know how the positive and negative instances are distributed in the citation space. Experimentation with several learning algorithms allows for a better understanding of the problem being addressed.
We propose to collect indexing results based on machine learning and MTI experiments, and use them as input data for the meta-learning experiments. The representation of the citation will play a role in the model optimization as well. For instance, n-grams afford an appropriate representation, when word collocation is relevant for indexing.
With small sets, manual selection and optimization of the parameters can be managed efficiently. But when
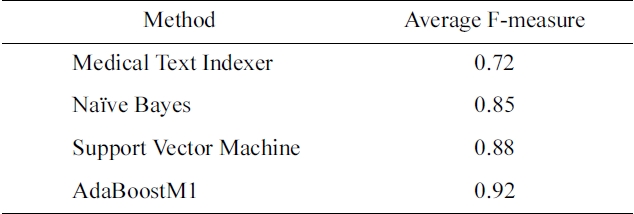
[Table 1.] F-measure for indexing methods on the Humans MeSH heading
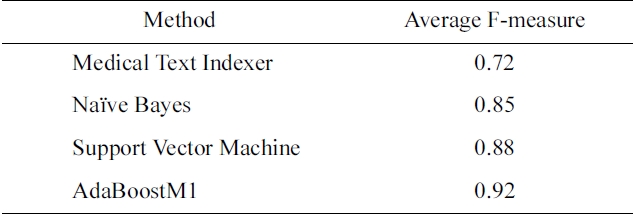
F-measure for indexing methods on the Humans MeSH heading
there is a large number of categories, meta-learning can play an important role. The optimization parameters are one level above traditional machine learning, since the objective is not to improve an existing learning algorithm, but to select the best algorithm, and its configuration for a given problem. In Table 1, we compare the performance of MTI and several standard machine learning algorithms for the Humans MeSH heading.
In this case, AdaBoostM1 outperforms all the other methods, and would be the method of choice for indexing citations with Humans MeSH heading.
In this section, we present how the framework is trained, and how it is used to index citations. Then, the base methods used for MeSH indexing are shown. The methods include MTI, a dictionary lookup approach, and several machine learning algorithms.
Experiments have been performed on a set of 300k citations from the 2011 MEDLINE Baseline and the 2011 MeSH vocabulary. The citations are sorted by date, so the first 200k citations are used for training, and the remaining 100k for testing.
The outcome of the training is a mapping between a MeSH heading, and an indexing method to be used for that MeSH heading. The performance of each algorithm on each MeSH heading was collected and compared. In this work, we have used the F-measure as our indexing performance measurement, which is standard in text categorization, even though other measurements, like accuracy, could be considered as well.
Since machine learning algorithms require training, we have split the 200k training data into training and validation subsets. To increase confidence, several training and validation splits were evaluated, and the results averaged. We have run 5 times 2-fold cross validation. Statistical significance of the results was computed using a randomization version of the two sample
In each split, the steps to estimate the performance of each algorithm A for each MeSH heading M were:
Step 1: If required, train algorithm A using the training subset. The positive examples are the citations indexed with the M MeSH heading, the rest are considered as negative examples. Note that MTI and dictionary lookup methods do not require training.
Step 2: Use algorithm A to index the citations in the validation subset with M MeSH heading.
Step 3: Compute the performance of algorithm A, i.e., the F-measure, comparing the indexing produced in step 2 to the original indexing for the validation set.
This process was repeated for each MeSH heading. The best method for each heading was selected and stored in a mapping table. For machine learning methods, the trained model for the best method was also stored in the table.
During indexing time, the mapping table prepared during the training process is used to index citations. Given a new citation, for each MeSH heading M the corresponding method from the mapping table is selected, and used to determine if the citation should be indexed with M.
Several implementations could be considered to speed up the indexing. Batch indexing of the citations, and a post-processing of the outcome, could be considered to index the citations with predictions by MTI, filtering out the predictions for which MTI was not the preferred method. On the other hand, trained machine learning models could be applied in parallel, to determine the indexing. This would allow processing of a large number of citations with one method, instead of processing a single citation by all the methods. Again, the results would be post-processed, but this time to merge the results of each indexing method.
Most of the indexing algorithms we study here require a training phase, MTI and dictionary lookup being exceptions. MTI has already been described in the introduction, so we focus on the other methods used in our experiments.
Since the main focus of the paper is the meta-learning framework, only machine learning algorithms that we could train using a large number of examples and a large number of categories (MeSH headings) have been selected. AdaBoostM1 has been used only in the Check Tag experiments. We are planning to include more learning algorithms, as they are integrated into our system.
This method looks for mentions of the MeSH heading in the citation text, as they appear in MeSH. If the mention of a MeSH heading is matched in the citation text, the citation is indexed with this MeSH heading. The preferred term and its entry terms are included in the dictio-nary. MeSH is turned into a list of terms and IDs.
Our dictionary lookup implementation is based on the monq.JFA package [23]. In addition to matching the dictionary terms to text, morphological changes are applied to the lexical items; e.g., the case of the first letter is normalized, hyphens are changed to spaces, and plural termination is normalized. Furthermore, the longest matched span is selected. For instance, the span of text “...quality of breast cancer care...” matches cancer and breast cancer. In this case, the match breast cancer is selected.
In our work, dictionary lookup was used to index a citation based on the title and abstract text (MeSH TIAB DL), and to index only the title (MeSH TI DL), which might provide higher precision, at the cost of recall.
A citation C is indexed with a MeSH heading
P(I|C)>P(NI|C) (1)
Using Bayes:
We can remove
As presented in Equation 3, the probability of a citation being indexed with a given MeSH heading is the product of the probabilities of each term
The probability of a term given a MeSH heading is estimated as shown in Equation 4, where
We use a smoothed model based on Jelinek-Mercer [24], due to term sparsity. In our experiments, we have used a value for λ of 0.8.
Finally, the prior
We have also implemented a variant of Naive Bayes (NB) based on term frequency - inverse document frequency (TF-IDF) [25], which has been shown to improve the performance of a traditional NB for text categorization.
We represent occurrences of the terms in the citations as binary features, so the frequency of a term in a document is not considered. We use a unigram model, so the relations of the terms in the citation are also not considered.
Usually used in query expansion in ad-hoc retrieval, Rocchio has been used as well for text categorization. A vector is calculated for each MeSH heading, by adding the mentions of the term
Given a citation, MeSH headings are ranked by cosine similarity. From this ranked list, we take the top n MeSH headings. In our experiments, we have considered the top 20.
AdaBoostM1 [26] is an ensemble learning algorithm, which samples iteratively from the training data, according to the performance of a base learner. In each iteration, a new model is produced. The final decision is based on the weighted sum of the models produced in the iterative process. The weights are estimated based on the performance of each model on the training data. In this work, 10 iterations were performed.
In our experiments, we used C4.5 as the base learner, since it has produced good results in the past [18], with a smaller set of MeSH headings. Our decision tree is an implementation of the C4.5 algorithm [27] with pruning, and with the minimum number of elements in leaf nodes set to 5. In our implementation, we consider binary features and 1-versus-all classification as well. This setup allows for optimizations in the information gain calculation that allow training this algorithm efficiently. We trained the learner on the random training set splits, as well as with oversampling of the positive examples, trying to overcome skewness in the distribution of positive and negative examples. In oversampling, examples are added to the minority category. In our experiments, we selected examples randomly from the minority category, till both categories had the same number of examples.
Combinations of methods have proved to increase performance of individual methods [28, 29]. Given a citation, for a given MeSH heading, the predictions for each of the indexing methods presented above were collected. Then, the votes were counted, and if the sum of the votes was over a given threshold, the MeSH heading was predicted by this method. We have performed experiments with different voting values, based on the methods presented above.
We have performed two experiments. In the first one, we have considered all the MeSH headings and trained algorithms, which can handle a large number of categories and features. In the second one, we evaluated a reduced set of MeSH headings, named check tags.
In both experiments, MTI annotation was considered the baseline method. Features for the machine learning algorithms were represented as the presence of tokens from the title and abstract of the citation; the frequency of the tokens in the citation was not used. The tokens were lowercased, but not stemmed.
>
A. Results with All MeSH Headings
This experiment was done on all MeSH headings. The
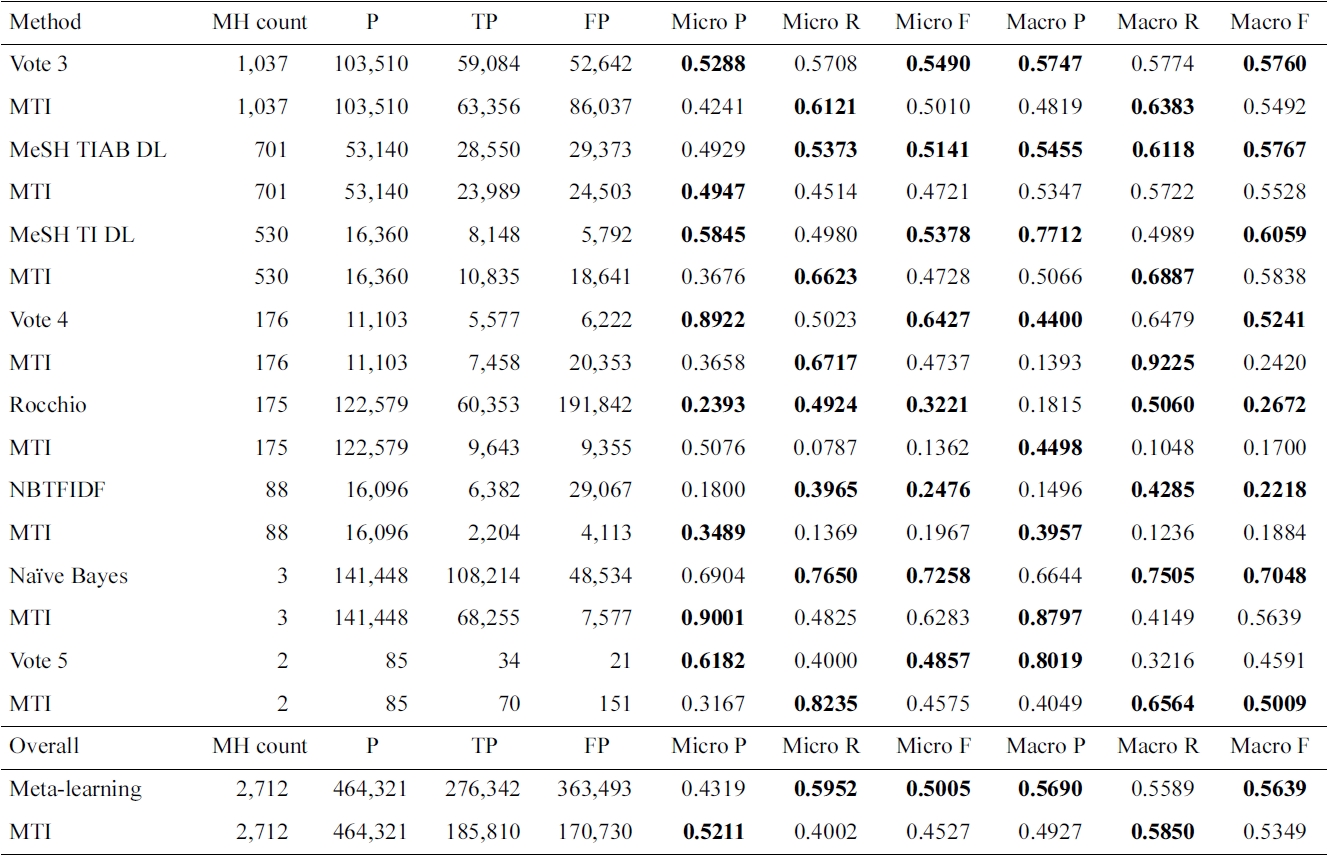
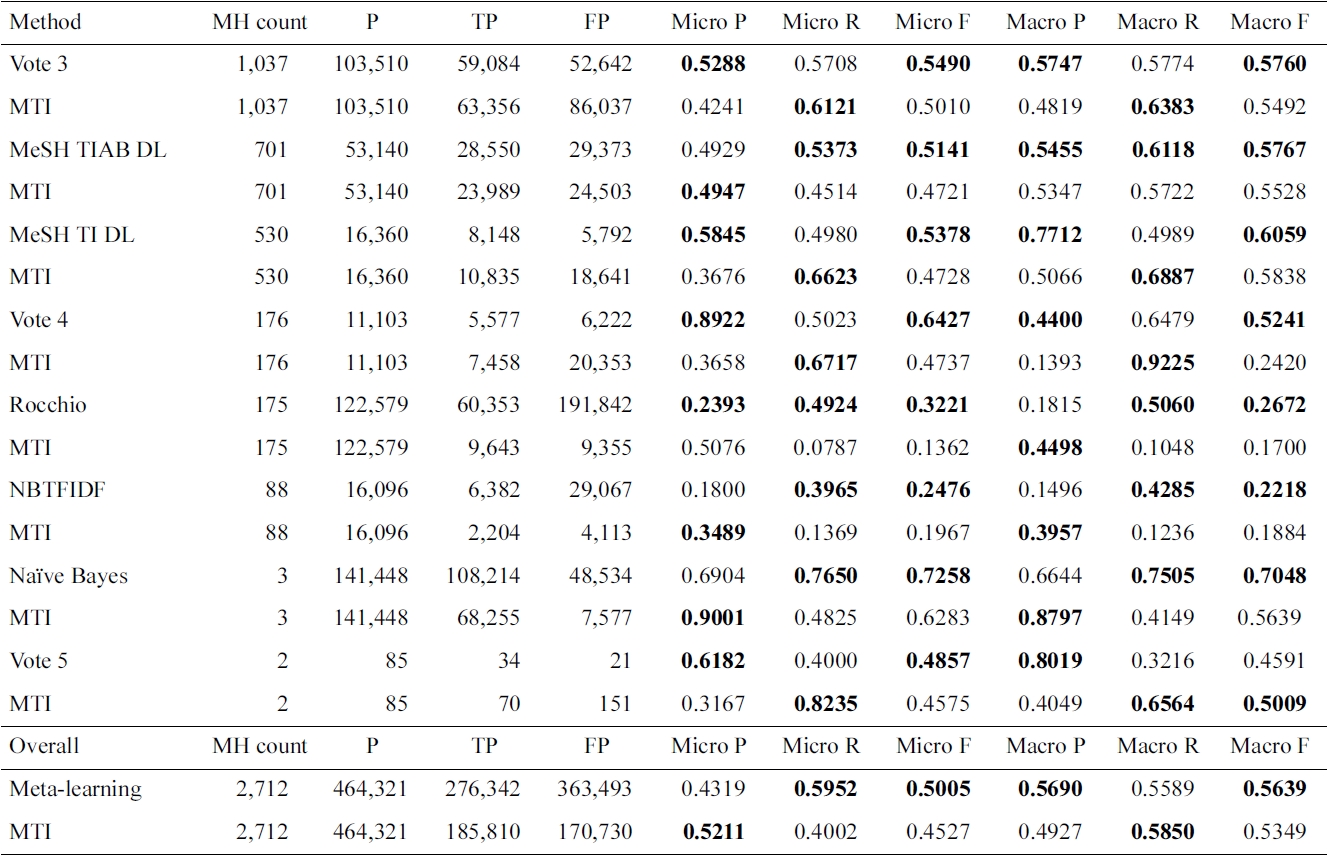
Results for Medical Text Indexer (MTI) and meta-learning for the 2,712 MeSH headings (MHs)
experiment used all but the AdaBoostM1 method, due to the time it takes to train it. For 2,712 of the 26k MeSH headings, a different method from MTI was selected: either a single method or a voting combination of them. Only methods significantly better than MTI were selected. This means that if the methods had a similar performance, MTI was preferred. In Table 2, we only show the set of MeSH headings grouped by learning method, where MTI was outperformed by methods as selected by meta-learning. MTI is the best algorithm for the MeSH headings not reflected in this table.
Voting 3, in which at least three methods agreed on predicting the MeSH heading, seems to perform better than the individual methods tested in this work. Voting 4 has a larger increase in precision compared to the decrease in recall, so the F1 is higher compared to MTI. On the other hand, if five of the evaluated methods need to agree, only a small number of MeSH headings are affected, and the performance is lower, compared to MTI.
Surprisingly, dictionary lookup (MeSH TIAB DL) performs reasonably well in some cases, compared to MTI. Machine learning methods perform better only on a small set of MeSH headings; one of the problems could be the small number of positive examples available for most of the MeSH headings.
Learning methods seemed to be more effective for the fewer MeSH headings that are more common in the indexing (e.g., Humans, Male, Female). These headings have more training data, and a more balanced proportion between positives and negatives.
NB has good performance on the most frequent MeSH headings (Humans, Male, and Female), which belong to the set of Check Tags. The modified NB (NBTFIDF) has better performance for a larger number of MeSH headings, compared to plain Naive Bayes. Finally, Rocchio performs better in a larger number of MeSH headings, compared to the other two NB algorithms.
We can see that only a limited number of MeSH head
[Table 3.] Results for Medical Text Indexer (MTI) and meta-learning for the Check Tags set
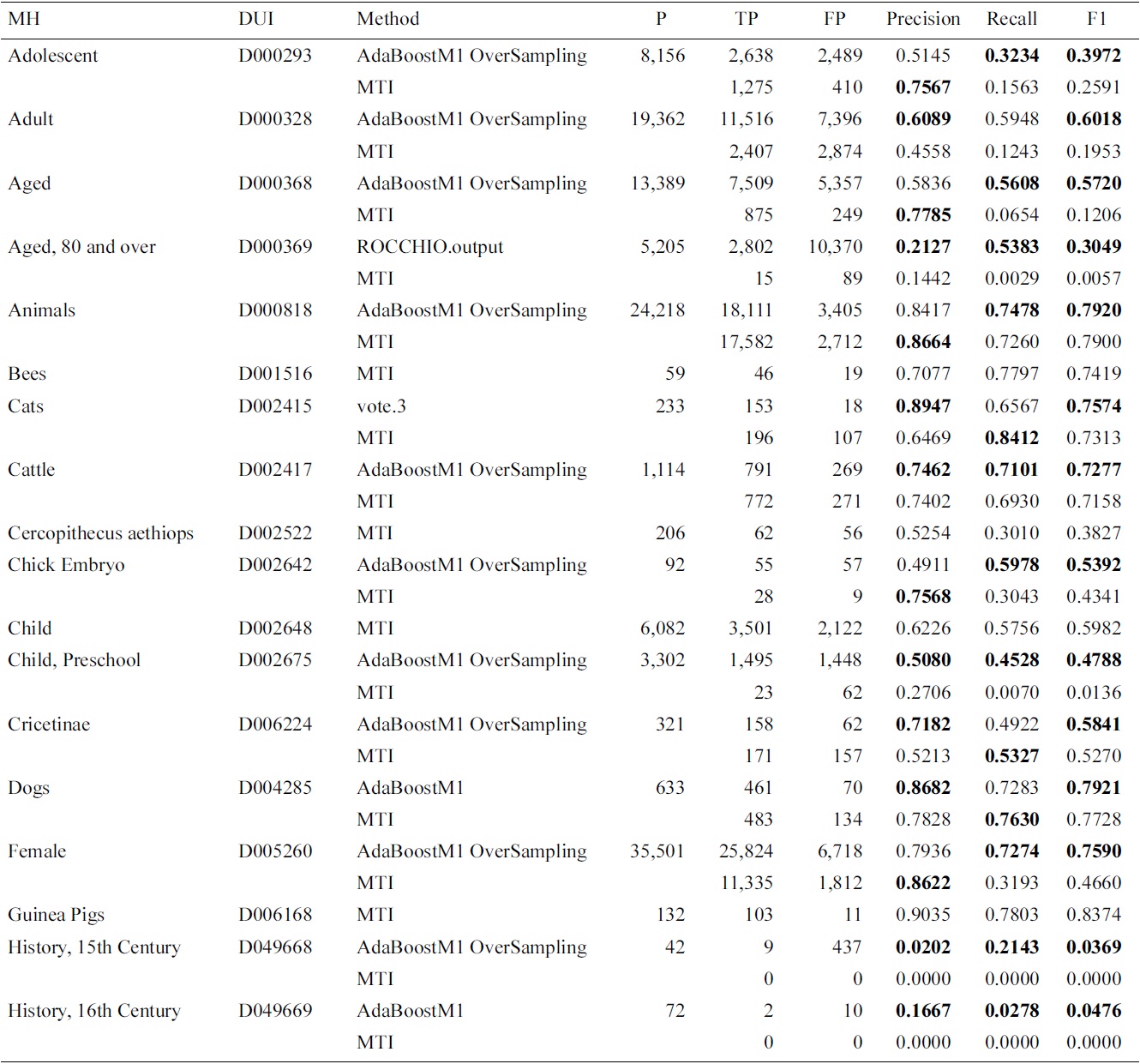
Results for Medical Text Indexer (MTI) and meta-learning for the Check Tags set
ings were affected by using the proposed approach. We have analyzed the results, and found that the improvements apply mostly to MeSH headings which had a higher indexing frequency. The large imbalance and variability between the training and testing might justify the results obtained with lower frequency MeSH headings. Another factor is that, since MTI is the current system, it has been left as the default, if the differences with MTI were not statistically significant.
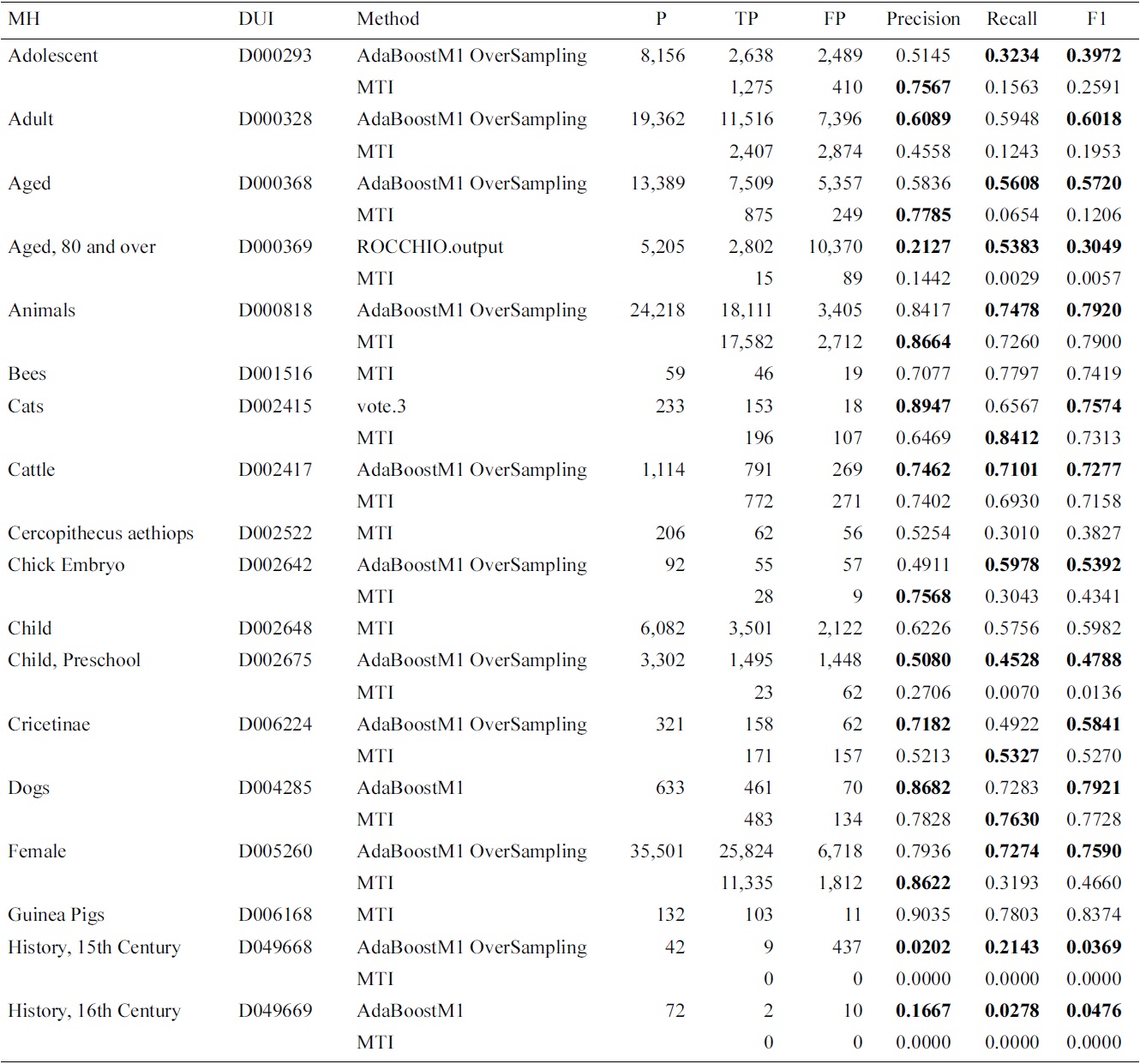
In this experiment, we have included AdaBoostM1 as a learning algorithm. In Table 3, we present the evalua-
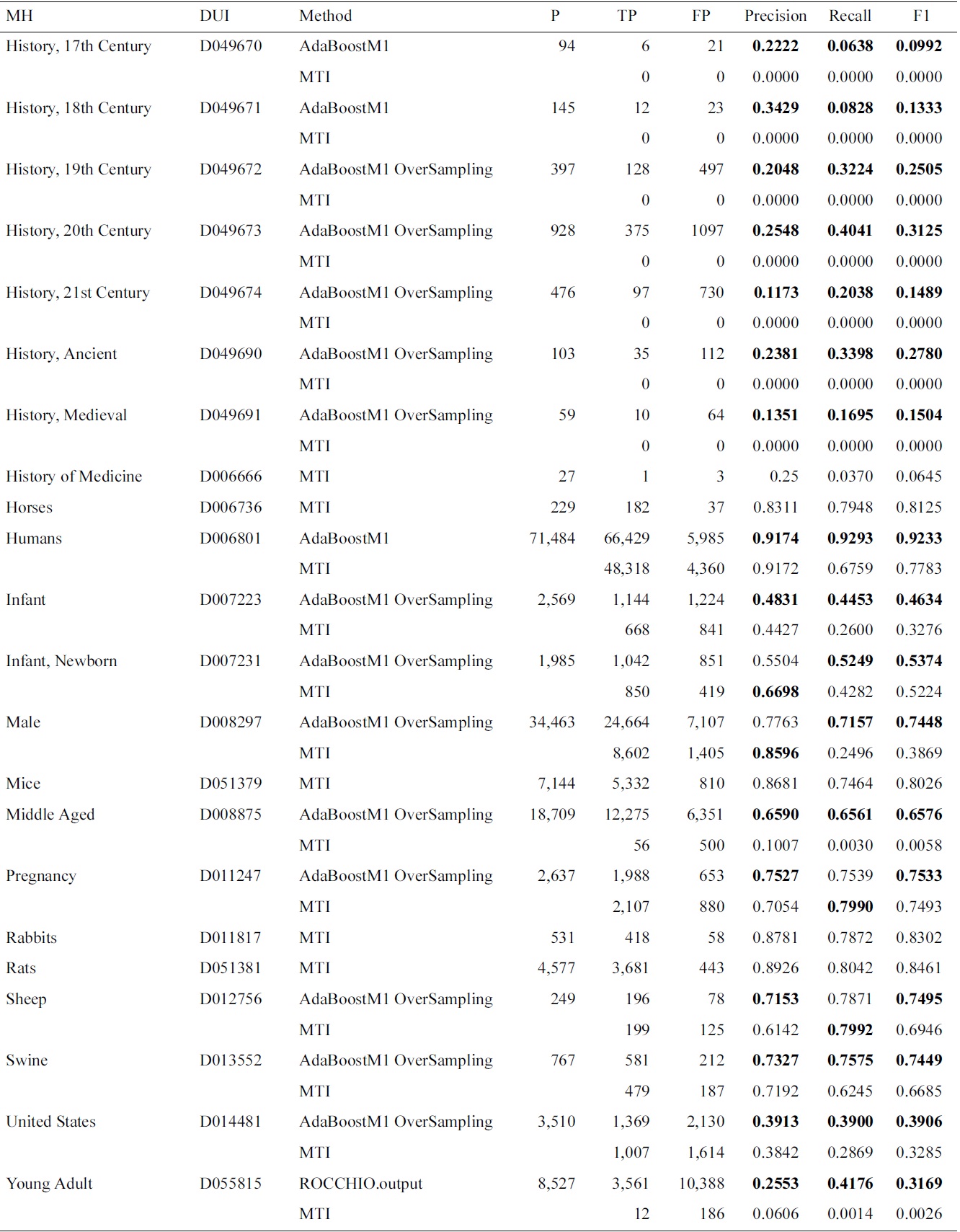
Continued
[Table 4.] Micro and macro results for the Check Tags set
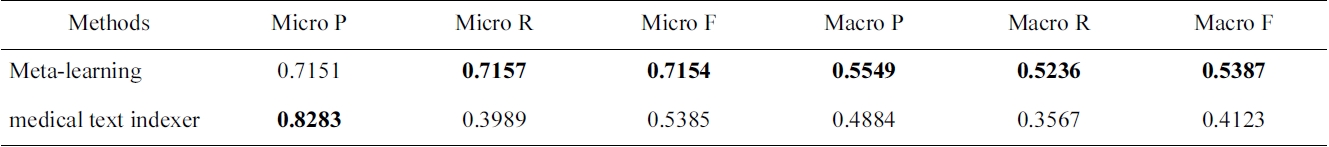
Micro and macro results for the Check Tags set
tion results for the selected method on the test data. We show that in most of the cases, the AdaBoostM1 with oversampling is the selected method. In Table 4, we compare overall Check Tag results with the MTI results. The performance of MTI is largely improved by meta-learning methods. In particular, Middle Aged, Young Adult and history-related terms profit from the use of alternative methods, which have very low MTI performance.
These results are in agreement with the experiments performed with all of MEDLINE, in which high frequency MeSH headings show a larger improvement, based on meta-learning.
VI. CONCLUSIONS AND FUTURE WORK
We have presented a framework that allows comparison of alternative indexing strategies, and an automated way of deciding on an optimal strategy for use with a large scale categorizer, namely MTI. We plan to add classifiers like SVMs, and to experiment with a larger set of MeSH headings with AdaBoostM1. In addition, we would like to include techniques that could learn with very imbalanced datasets, to improve the performance in lower frequency MeSH headings. The software used for these experiments is available at [30].
We have considered only the text from the title and abstract. More information is available in MEDLINE metadata, which might be exploited; examples include the journal and author affiliations.
Other sampling techniques, like synthetic sampling, might overcome some of the problems of oversampling and undersampling.
We would like to work as well on the automatic combination of indexing methods. This may require a combination of features and models, in which genetic programming might play a relevant role.