The importance of illustrations in scientific publications is well-established. In a survey of information needs of researchers and educators, Sandusky and Tenopir [1] found that scientific journal-article components, such as tables and figures, are often among the first parts of an article scanned or read by researchers. In addition, the survey participants indicated that having access to the illustrations (we use “images,” “illustrations,” and “figures” interchangeably when referring to visual material in our set of medical articles) prior to obtaining the whole publication would greatly enhance their search experience. In the biomedical domain, Divoli et al. [2] found that bioscience literature search systems, such as PubMed, should show figures from articles alongside search results, and that captions should be searched, along with the article title, metadata, and abstract. Simpson et al. [3] showed that, for the system presented in this paper, retrieval of case descriptions similar to a patient’s case was significantly improved with the use of image-related text.
The clear need for a multimodal retrieval system on the one hand, and the sufficient maturity of the information retrieval (IR) and content-based image retrieval (CBIR) techniques on the other, motivated us to implement a prototype multimodal system (called OpenI) for advanced information services. These services should enable:
Searching by textual, visual and hybrid queries
Retrieving illustrations (medical images, charts, graphs, diagrams, and other figures)
Retrieving bibliographic citations, enriched by relevant images
Retrieving from collections of journal literature, patient records, and independent image databases
Linking patient records to literature and image databases, to support visual diagnosis and clinical decision making
This paper first presents an overview of the processes involved in preparing multimodal scientific articles for indexing and retrieval. It then briefly introduces a distributed architecture that allows for both processing the original documents in reasonable time (for example, extracting image features from thousands of images in just a few hours), and for real-time retrieval of the processed documents. The paper concludes with a discussion of the implemented system prototype (Fig. 1) that currently provides access to over 600,000 figures from over 250,000 medical articles, evaluation of the algorithms constituting the system, and future directions in multimodal retrieval and its evaluation.
II. BUILDING BLOCKS FOR ADVANCED INFORMATION SERVICES
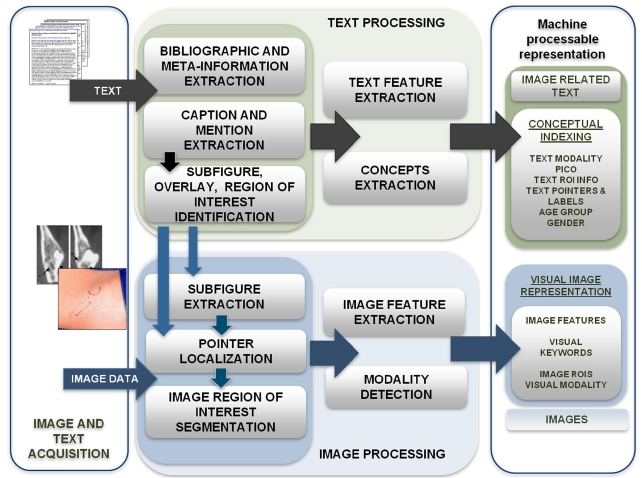
To prepare documents for indexing and retrieval, we combine our tools and those publicly available, in a pipeline that starts with acquiring data and ends in the generation of MEDLINE citations enriched with image-related information (henceforth, “enriched citations”). The initially separate text and image processing pathways merge to create multimodal indexes, for use with specialized mul-
timodal information retrieval algorithms in Fig. 2. The images and text data in the current prototype are obtained from the open access subset of PubMed Central (PMC), using the PMC file transfer protocol (FTP) services (http://www.ncbi.nlm.nih.gov/pmc/tools/openftlist/). This full-text archive of biomedical publications provides the text of each article in extensible markup language (XML) format, and all published figures as JPEG files. The XML files serve as input to the modules assembled in the text processing pipeline, and the images are processed through the image processing pipeline. Several image processing modules (for example, modality classifiers) require output of the text processing modules as additional input.
The output of the document processing pipeline is a set of enriched MEDLINE citations in XML format that is subsequently indexed with the National Library of Medicine (NLM)’s domain-specific search engine Essie [4], as well as with the widely-used open-source search engine Lucene (http://lucene.apache.org/). An enriched citation consists of three parts: 1) the original bibliographic citation obtained using E-Utilities (http://www.ncbi.nlm.nih.gov/corehtml/ query/static/eutils_help.html); 2) the image caption and image-related paragraphs extracted from the full-text of the article, along with salient information extracted from this free text and stored in structured form; and 3) image features expressed as searchable alphanumeric strings, along with the image uniform resource identifier (URI), for display in the user interface.
Some of the processing steps (such as extracting elements of an XML document) are well-known, and will be omitted here. We will focus instead on the overall process flow, and the unique challenges and opportunities presented by images found in biomedical publications.
>
A. Generating an Enriched Citation
The OpenI document processing system is developed in Java, and uses Hadoop MapReduce (Apache Software Foundation, Los Angeles, CA, USA) to parallelize text processing and image feature extraction. An enriched citation object is generated in the text processing pipeline, which is presented in Section III. Images are processed independently, and the information extracted from the images is added to the enriched citation in the final merging step.
One challenge in image processing arises from several illustrations combined into one figure. These multi-panel (or compound) images found in many articles reduce the quality of image features, if the features are extracted from the whole image. For feature extraction, therefore, these images need to be first separated into distinct panels. This process is described in Section V.
In addition to the text and image features necessary for retrieval, each enriched citation also contains meta-information derived from the basic features (such as the medical terms found in the captions and mapped to the unified medical language system [UMLS] [5] concepts). This meta-information is used to filter and re-rank search results. For example, the results could be restricted to radiology images using the modality classification results, or reranked to promote articles focused on genetics (identified as such by genetics-related concepts in the titles, Medical Subject Heading [MeSH] terms, and captions). The currently available filters are described in Section VI.
The text processing begins with the extraction of the image caption and the paragraph(s) discussing the figure (“mentions”). In the PMC documents, captions are a defined XML element. We extract the mentions using regular expressions: we first find an indicator that a figure is mentioned, usually, words “Figure” or “Fig” (sometimes within mark-up tags or punctuation) followed by a number, and then extract the paragraph around the indicator.
Next, the caption processing module determines if the caption belongs to a multi-panel figure. The rule-based system is looking for sequences of alphanumeric characters that are included within repeating tags, or followed by a repeating punctuation sign (for example, A. B. C.) If a sequence is found, the number of panels and the panel labels are added to the enriched citation.
The next module extracts the descriptions of image overlays (such as arrows), and regions of interest (ROI) indicated by the overlays [6]. The ROI descriptions added to the enriched citations are currently a searchable field. The whole output of the module is needed for our ongoing research, in building a visual ontology that will associate the UMLS concepts with specific image features.
Finally, a concept extraction module submits the captions and mentions to MetaMap [7], which identifies UMLS concepts in the text. The module then applies rules and stop-word lists to the MetaMap output, to reduce the set of the identified UMLS concepts to the salient disorder, intervention and anatomy terms [8]. Fig. 3 shows an enriched citation in XML format.
Low-level visual features, such as color, texture, and shape, are insufficient for capturing image semantics, but they are the primary building blocks of the visual content in an image. They can be effective, if a judiciously selected feature metric is used to capture the visual content in an image, and then incorporates it into a suitable machinelearning framework that supports multi-scalar and concept- sensitive visual similarity. In the OpenI prototype system, the low-level visual features extracted from the whole set of images are first clustered using the k-means algorithm, and then the resulting clusters are labeled with alphanumeric strings (“cluster words”) that serve as cluster identifiers. The images are then annotated with the cluster identifier of each low-level feature. These cluster words are added to the enriched citation in Fig. 3, as the last document preparation step. The enriched citation field
containing cluster words is indexed in the same manner as the rest of the citation.
Images in the open access PMC collection are of different sizes. In order to obtain a uniform measure with greater computational efficiency, we compute features from images that are reduced to a common size, measuring 256 × 256 pixels. In the future, we intend to process images at a significantly higher (or full) resolution, to extract meaningful local features.
1) Color Features
Color plays an important role in the human visual system, and measuring its distribution can provide valuable discriminating data about the image. We use several color descriptors to represent the color in the image. To represent the spatial structure of images, we utilize the color layout descriptor (CLD) [9] specified by MPEG-7 [10]. The CLD represents the spatial layout of the images in a compact form, and can be computed by applying the discrete cosine transformation (DCT) to the 2D array of local representative colors in the YCbCr color space, where Y is the luminance component, and Cb and Cr are the blue and red chrominance components, respectively. Each color channel is 8-bits, and is represented by an average value computed over 8 × 8 image blocks. We extract a CLD with 10 Y, 3 Cb, and 3 Cr components, to form a 16-dimensional feature vector.
Another feature used is the color coherence vector (CCV) [11] that captures the degree to which pixels of that color are members of large similarly colored regions. A CCV stores the number of coherent versus incoherent pixels with each color, thereby providing finer distinctions than color histograms. Color moments, also computed in the perceptually linear L*a*b* color space, are measured, using the three central color moment features: mean, standard deviation, and skewness. Finally, 4 dominant colors in the standard red, green, blue (RGB) color space and their degrees are computed, using the k-means clustering algorithm.
2) Edge Features
Edges are not only useful in determining object outlines, but their overall layout can be useful in discriminating between images. The edge histogram descriptor (EHD) [9], also specified by MPEG-7, represents a spatial distribution of edges in an image. It computes local edge distributions in an image, by dividing the image into 4 × 4 sub-images, and generating a coarse-orientation histogram from the edges present in each of these subimages. Edges in the image are categorized into five types: vertical, horizontal, 45 × diagonal, 135 × diagonal, and other non-directional edges. A finer-grained histogram of edge directions (72 bins of 5 × each) is also constructed from the output of a Canny edge detection algorithm [12] operating on the image. This feature is made invariant to image scale, by normalizing it with respect to the number of edge points in the image.
3) Texture Features
Texture measures the degree of “smoothness” (or “roughness”) in an image. We extract texture features from the four directional gray-level co-occurrence matrices (GLCM) that are computed over an image. Normalized GLCMs are used to compute higher order features, such as energy, entropy, contrast, homogeneity and maximum probability. We also compute Gabor filters to capture image gist (coarse texture and spatial layout). The gist computation is resistant to image degradation, and has been shown to be very effective for natural scene images [13]. Finally, we use the discrete wavelet transform (DWT) that has been shown to be useful in multiresolution image analysis. It captures image spatial frequency components at varying scales. We compute the mean and standard deviation of the magnitude of the vertical, horizontal, and diagonal frequencies at three scales.
4) Average Gray Level Feature
This feature is extracted from the low-resolution scaled images, where each image is converted to an 8-bit graylevel image, and scaled down to 64 × 64 pixels, regardless of the original aspect ratio. Next, this reduced image is partitioned further with a 16 × 16 grid, to form small blocks of (4 × 4) pixels. The average gray value of each block is measured and concatenated, to form a 256- dimensional feature vector.
5) Other Features
We extract two additional features using the Lucene image retrieval engine (LIRE) library: the color edge direction descriptor (CEDD) and the fuzzy color texture histogram (FCTH) [14]. CEDD incorporates color and texture information into a single histogram, and requires low computational power compared to MPEG-7 descriptors. To extract texture information, CEDD uses a fuzzy version of the five directional edge filters used in MPEG- 7 EHD that were described previously. This descriptor is robust with respect to image deformation, noise, and smoothing. The FCTH uses fuzzy high frequency bands of the Haar wavelet transform to extract image texture.
V. MULTI-PANEL FIGURE SEGMENTATION
Independent of the caption processing module that outputs the number of panels and panel labels, two imagefeature based modules detect panel boundaries and panel labels. The output of the three modules is used in the panel splitting module.
The image-feature based panel segmentation module determines if an image contains homogeneous regions that cross the entire image. If no homogeneous regions are found, the image is classified as single-panel. If homogeneous regions are found, the panel segmentation module iteratively determines if each panel contains homogenous regions, and finally outputs coordinates of each panel.
The label detection module [15] first binarizes the image into black and white pixels, then, searches the image for connected components that could represent panel labels, and then applies optical character recognition (OCR) methods to the connected components. Finally, the most probable label sequences and locations are selected from all candidate labels, using Markov random field modeling.
The label splitting module takes the outputs of the caption splitting, panel segmentation and label detection modules, and splits the original figure if the following conditions are met: all three modules agree on the number of panels, and the caption splitting and label detecting modules agree on the labels (this happens for approximately 30% of the multi-panel figures). If the panel segmentation or the label detection modules fail completely, the image cannot be split. However, if the modules partially agree on labels, and position some of the labels at the corners of the corresponding panels, heuristics help to compensate for the partial errors of individual modules, and the combined information helps correctly split another 40% of the multi-panel images. The images on which the algorithm fails are processed as single-panel images. All images are displayed in the original form.
OpenI uses VMware vSphere 4 (VMware Inc., Palo Alto, CA, USA) and a high-performance Linux based storage area network (SAN) to support a fault-tolerant, scalable, and efficient production-grade system. A highperformance SAN storage cluster that was built using Red Hat Enterprise Linux (Red Hat, Raleigh, NC, USA) provides OpenI with a dedicated, reliable, predictable and high-performing storage system for the Hadoop cluster. VMware vSphere 4 is used to virtualize the Hadoop Namenode, and run it in fault-tolerant mode. The faulttolerance feature of VMware allows a single virtual machine to run simultaneously on two hardware servers. Further, vSphere monitors the heartbeat of the Namenode, and restarts it automatically if it should become inoperable. These features eliminate the single point of failure, and turn Hadoop into a stable and reliable development and research platform. Finally, vSphere is used to make OpenI processes run efficiently on multi-core CPUs. Each compute node running processes that are not multicore aware can be created as a virtual machine tied to a specific physical core.
VII. OPENI IMAGE RETRIEVAL SYSTEM
The OpenI prototype (http://openi.nlm.nih.gov/) currently supports image retrieval for textual, visual and hybrid queries. The images submitted as queries are processed as described above, and represented using cluster “words”. After this processing step, the cluster “words” are treated as any other search terms.
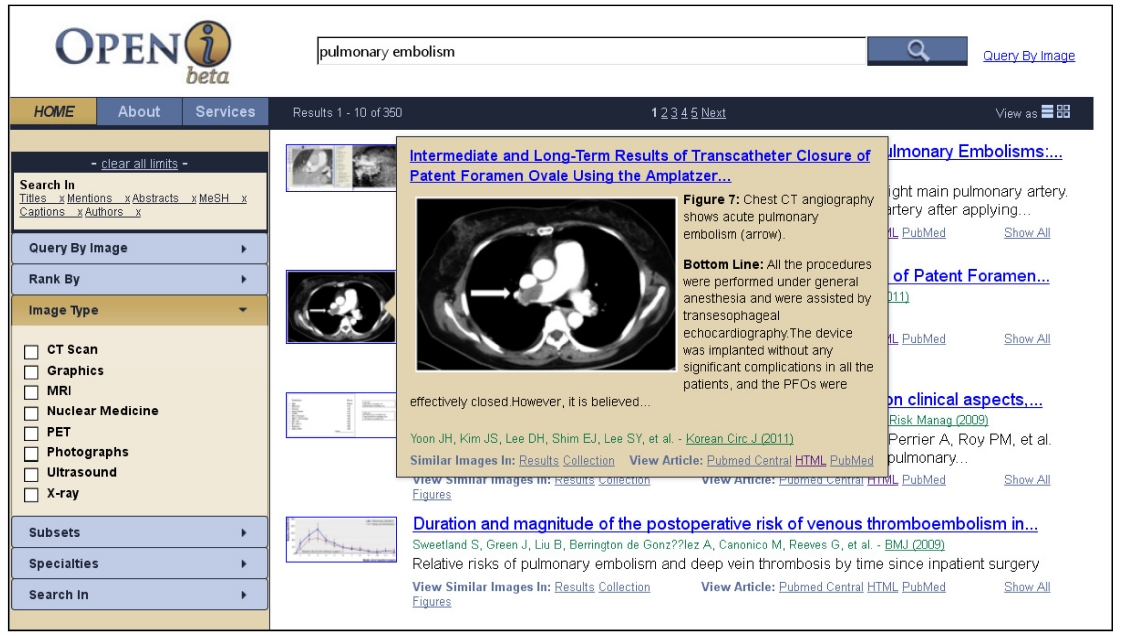
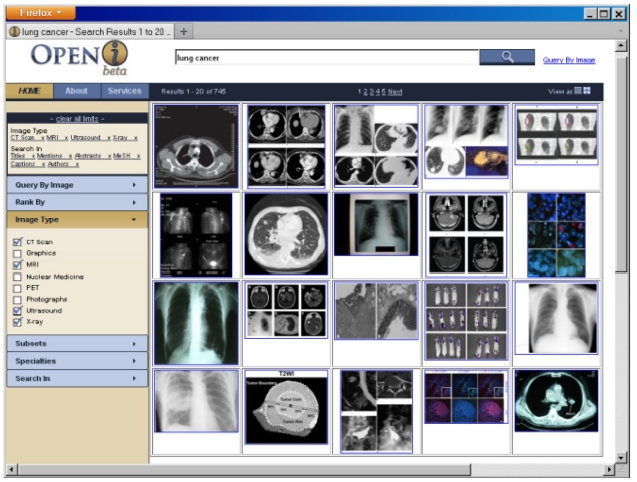
Based on the principles developed by [16], the search results are displayed either on a grid that allows a view of all top 20 retrieved images (Fig. 4), or as a traditional list in Fig. 1. In either layout, scrolling over the image brings
up a pop-up window that, along with the traditional elements of search results, such as titles and author names, provides captions of the retrieved images, and short summaries of the articles.
The summaries are obtained through RIDeM services (http://clinicalreferences.nlm.nih.gov/ridem). The summaries (or “bottom-line” patient-oriented outcomes extracted from abstracts) are generated by extracting three sentences most likely to discuss patient-oriented outcomes of the methods presented in the paper [17]. The probability of a sentence to be an outcome is determined by a metaclassifier that combines outputs of several base classifiers (such as a Na?ve Bayes classifier, and classifiers based on the position of the sentence in the abstract, or the presence of relevant terms in the sentence).
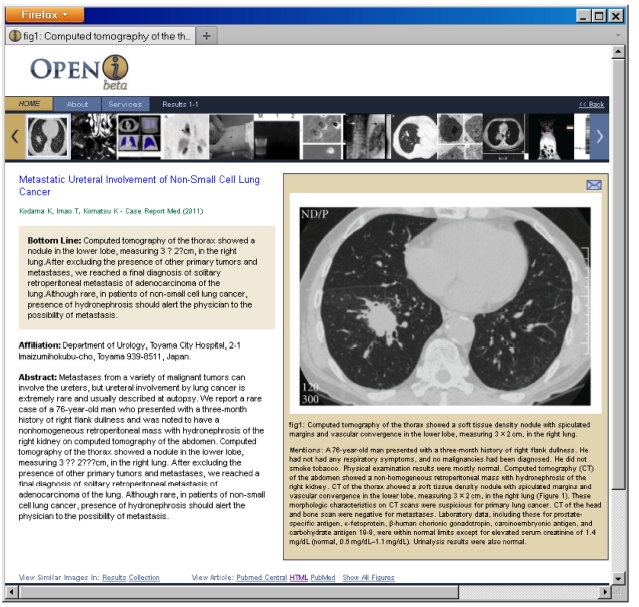
Once the search results are displayed, the users can find similar images in the results and in the entire collection, by following links that perform new search requests for images visually similar to the currently selected image. These new searches are based purely on image features. Users may view all other images in a given paper without leaving the initial search page, and drill down to the full enriched citation in Fig. 5.
The search results can also be filtered using the following facets: 1) image type; 2) subsets of publications, for example, systematic reviews; 3) clinical specialties; 4) enriched citation fields.
The image type filter is based on our classification of images into eight medical images modalities, such as magnetic resonance imaging (MRI), x-ray, computed tomography (CT), and ultrasound. Our method [18] uses a support vector machine (SVM) to classify images into multiple modality categories. The degree of membership in each category can then be used to compute the image modality. In its basic formulation, the SVM is a binary classification method that constructs a decision surface, and maximizes the inter-class boundary between the samples. To extend it to multi-class classification, we combine all pair-wise comparisons of binary SVM classifiers, known as one-against-one or pair-wise coupling (PWC). Each SVM is trained for one image feature. The class with the greatest estimated probability for each feature accumulates one vote. The class with the greatest number of votes after classifying for all features is deemed to be the winning class, and the modality category of the class is assigned to the image. When a user requests a specific image type, a hard constraint on exactly matching the image modality field of the enriched citation is imposed.
Due to the nature of the collection, not all MEDLINE/ PubMed subsets (http://www.nlm.nih.gov/bsd/pubmed_subsets. html), such as the core clinical journals subset (http://www.nlm.nih.gov/bsd/aim.html), are available in OpenI. We used the subject field of the NLM’s List of Serials Indexed for Online Users (http://wwwcf.nlm.nih.gov/ serials/journals/) to categorize the journals into clinical specialties and subsets. Not all journals are assigned to broad subject areas, and only publications in journals that are assigned to an area will be retrieved, when a user requests to filter the results by a subset or specialty. Where available, we used the subset field of the original MEDLINE citation.
The users can search the text in any combinations of the following: titles, abstracts, captions, mentions, MeSH terms, and author names.
Finally, the search results could also be re-ranked according to the users’ interests (indicated by selecting advanced search options) along the following axes: 1) by the date of publication (most recent or oldest first); 2) by the clinical task that is discussed in the paper (diagnosis, cause of the problem, prevention, prognosis, treatment, etc.). The clinical task that was the focus of the study presented in a paper is determined using rules that take into account specific MeSH terms. For example, the term “Infectious Disease Transmission, Vertical/*Prevention & Control” indicates that the task is prevention. The star indicates that the publication is focused on prevention.
Several ongoing research efforts are dedicated to augmenting text results with images. Some of these efforts aim to retrieve images by matching query text terms in the citations to the articles and the figure captions. We list five efforts related to our goals. Most systems do not use image features to find similar images or combine visual and text features for biomedical information retrieval. Our goals include improving the relevance of multi-modal (text and image) information retrieval, by including lessons learned from these efforts.
The BioText [16] search engine searches over 300 open access journals, and retrieves figures, as well as text. Bio- Text uses Lucene to search full-text or abstracts of journal articles, as well as image and table captions. Retrieved results (displayed in a list or grid view) can be sorted by date or relevance. This search engine has influenced our user interface design.
Yottalook (http://www.yottalook.com/) allows multilingual searching to retrieve information (text or medical images) from the Web and journal articles. The goal of the search engine is to provide information to clinicians at the point of care. The results can be viewed as thumbnails or details. This site sets an example in the breadth of its searches, capabilities to filter results on image modality and other criteria, being current with social media, and connecting with the users’ myRSNA accounts (offered by the Radiological Society of North America [RSNA]), which allow saving of search results.
Other related work includes the Goldminer (http://goldminer. arrs.org/home.php) search engine developed by the American Roentgen Ray Society (ARRS) that retrieves images by searching figure captions in the peer-reviewed journal articles appearing in the RSNA journals, Radiographics and Radiology. It maps keywords in figure captions to concepts from the UMLS. Users have the options to search by age/modality/sex for images, where such information is available. Results are displayed in a list or grid view.
The FigureSearch (http://figuresearch.askhermes.org/articlesearch/ index.php?mode=figure) system uses a supervised machine-learning algorithm for classifying clinical questions, and Lucene for information retrieval over the published medical literature, to generate a list view of the results with relevant images, abstracts, and summaries.
The Yale image finder (YIF) [19] searches text within biomedical images, captions, abstracts, and titles, to retrieve images from biomedical journal papers. YIF uses optical character recognition to recognize text in images, in both landscape and portrait modes.
The image retrieval in medical applications (IRMA) system (http://www.irma-project.org) primarily uses visual features and a limited number of text labels that describe the anatomy, biosystem, imaging direction, and modality of the image for medical image retrieval. We have collaborated with the developers of the IRMA system, and enhanced their image retrieval system (which uses features computed on the gross image), with our image features and similarity computation techniques applied to local image regions [20].
Increasing commercial interest in multi-modal information retrieval in the biomedical domain is indicated by the industry teams participating in the ImageCLEFmed (http://www.imageclef.org/2012/medical) evaluations dedicated to retrieval of medical images and similar patients’ cases. Participants include researchers from Siemens, GE Medical Systems, Xerox, and other industrial organizations. Publishers such as Springer also provide text-based image retrieval (http://www.springerimages.com/). Other commercial image search engines include those developed by Google, Gazopa, and Flickr.
Saracevic [21] defines six levels of evaluation of information retrieval (IR) systems: 1) the engineering level addresses speed, integrity, flexibility, computational effectiveness, etc.; 2) the input level evaluates the document collection, indexed by the system and its coverage; 3) the processing level assesses performance of algorithms; 4) the output level evaluates retrieval results and interactions with the system; 5) the use and user level evaluates system performance for a given task; and 6) the social level evaluates the impact of the system on research, decision-making, etc.
OpenI is a complex system that has been evaluated along several of the aforementioned axes: on engineering, processing, and output levels. Its modality classification, ad-hoc image retrieval and case-based retrieval have been evaluated within the ImageCLEFmed evaluations since 2007. The OpenI results are steadily in the leading group, achieving, for example, 92% accuracy in the modality classification task in 2010, and 0.34 mean average precision in case-based retrieval in 2009.
In addition to testing the overall retrieval performance of the system, we have evaluated the benefits of enriching MEDLINE citations with image captions and passages pertaining to images [3]. As mentioned above, these passages significantly improved case-based retrieval. Other evaluations of the parts of the document preparation steps include evaluation of multi-panel image segmentation, and evaluation of the processes involved in automatic generation of a visual ontology, such as the ROI and ROI marker extraction from text and images. Overall, the panel splitting module achieves 80.92% precision at 73.39% recall. The ROI marker extraction from text achieves 93.64% precision and 87.69% recall, whereas ROI identification is at 61.15% accuracy [6]. The ROI marker extraction (limited to arrows) ranges from 75% to 87% accuracy for different arrow types [22].
User level evaluations are often approximated with site visits and click-through data [23]. To that end, we are monitoring the numbers of unique visitors per day, which for May 2012 is at approximately 700 on average, and shows a growing trend. We are also planning a social level experiment (with members of particular research and clinical communities) to evaluate the effectiveness of OpenI in assisting with specific tasks.
The deployment of the prototype system and the architecture presented in this paper allows continuing research in several directions. First, we are interested in the usability of the current user interface, and the usefulness of the search features. Second, we are expanding and improving the extraction of the basic image features, and the selection of these features for various higher-level tasks, such as image modality classification, image ROI recognition, and building a visual ontology. The latter task includes associating specific image features with the UMLS concepts. Recognizing that many researchers would like to focus on specific aspects of image retrieval, for example improving retrieval methods or text understanding, we plan to provide our current document processing methods as publicly available services.