Water is a major vehicle of disease transmission preceded scientific verification of the germ theory of disease [1]. Therefore, the presence of infectious bacteria, such as
Among the bacterial properties, viability and activity (bacterial regrowth potential, BRP) assessments are major requirements in several areas of infectious bacteriology [3]. Especially, detection of viability of pathogenic bacteria is very important to assess the inactivation profiles in the water disinfection process. Assessment of active or viable microorganisms is often difficult, since no single analytical method identifies all physiological characteristics of bacterium under a certain condition [4]. Although counting colony formed on agar plate is usually employed as the standard method for measuring bacterial viability, it only indicates the number of cells able to replicate under the conditions provided for growth. Moreover, the counting colony forming unit (CFU) method requires much time (in most cases, results are available in 24 to 72 hr) to incubate and detect the target cells, and some bacteria in a specific physiological status, for example in a viable but nonculturable state, cannot be detected by the CFU counting method [2, 5].
Fluorescent techniques in combination with flow cytometry (FCM) have been extensively used for the assessment of the viability or BRP of microorganisms from different environmental samples [5-7]. The FCM technique makes it possible to perform a rapid detection of individual cell viability within a few minutes [8-10], and offers a powerful tool for analyzing a cell population at the single-cell level, since it can be used both to identify and enumerate bacterial populations from environmental samples and to characterize functional properties of the individual cell [5, 10]. Moreover, it allows simultaneous measurement of different physical and biochemical parameters and hence offers substantial information on the dynamics and physiological heterogeneity of the bacterial population. In addition, FCM offers the ability to physically separate selected cells by cell sorting for further molecular and physiological analysis when used in combination with fluorescent staining, for example, dyes based on enzyme activity, membrane integrity, and membrane potential [11-16].
FCM is an analytical method that allows the rapid measurement of light scattered and fluorescence emission produced by suitably illuminated cells. The cells, or particles, are suspended in liquid and produce signals when they pass individually through a beam of light [6]. Since measurements of each particle or cell are made separately, the results represent cumulative individual cytometric characteristics. An important analytical feature of FCMs is their ability to measure multiple cellular parameters [5, 16]. Some FCMs are able to physically separate cell subsets (sorting) based on their cytometric characteristics.
However, most of the FCM tests that assess the viability of bacteria have been done certainly about the single sort of pure culture cell in laboratory environments, so far. Furthermore, the applicability of FCM to the viability assessment as well as the detection of the amount of dead bacteria with regards to the water disinfection field has not been clearly studied thus far.
Therefore, this study was performed to estimate the applicability of FCM to assess the viability as well as the population of
Two bacterial strains,
2.2. Preparation of LIVE and DEAD Bacteria
Three pairs of sterilized centrifuging tubes (15 mL) including 10 mL of each culture media (LB broth, CBYE) containing a number of
Additionally, silver and copper cations dissolved from silver nitrate (AgNO3; Kisita Co., Japan) and copper sulfate, 5-hydrate (CuSO4 ?5H2O; Kisita Co., Japan) was exposed to
2.3. Staining Protocols and FCM
Based on the permeability of the cell membrane, the nucleic acids (RNA/DNA) of
FCM analyses were performed on a EPICSⓡ ALTRA™ with a HyPerSort Cell Sorting flow cytometer (Beckman Coulter Inc.) equipped with a 15-mW, 488 nm, air cooled argon ion laser. Bacterial samples were diluted to approximately 105 cells/mL and delivered at a low flow rate, corresponding to 80 to 150 cells or particles/sec. A band pass filter of 525 nm (510 to 540 nm) was
used to collect the green fluorescent signal (PMT2), and a band pass filter of 610 nm (615 to 640 nm) was used to collect the red fluorescent signal (PMT4). Data were analyzed with the EXPO™ 2 Flow Cytometry Software ver. 2 (Beckman Coulter Inc.).
3.1. FCM Protocol for the Double Staining with SYTO 9 and PI
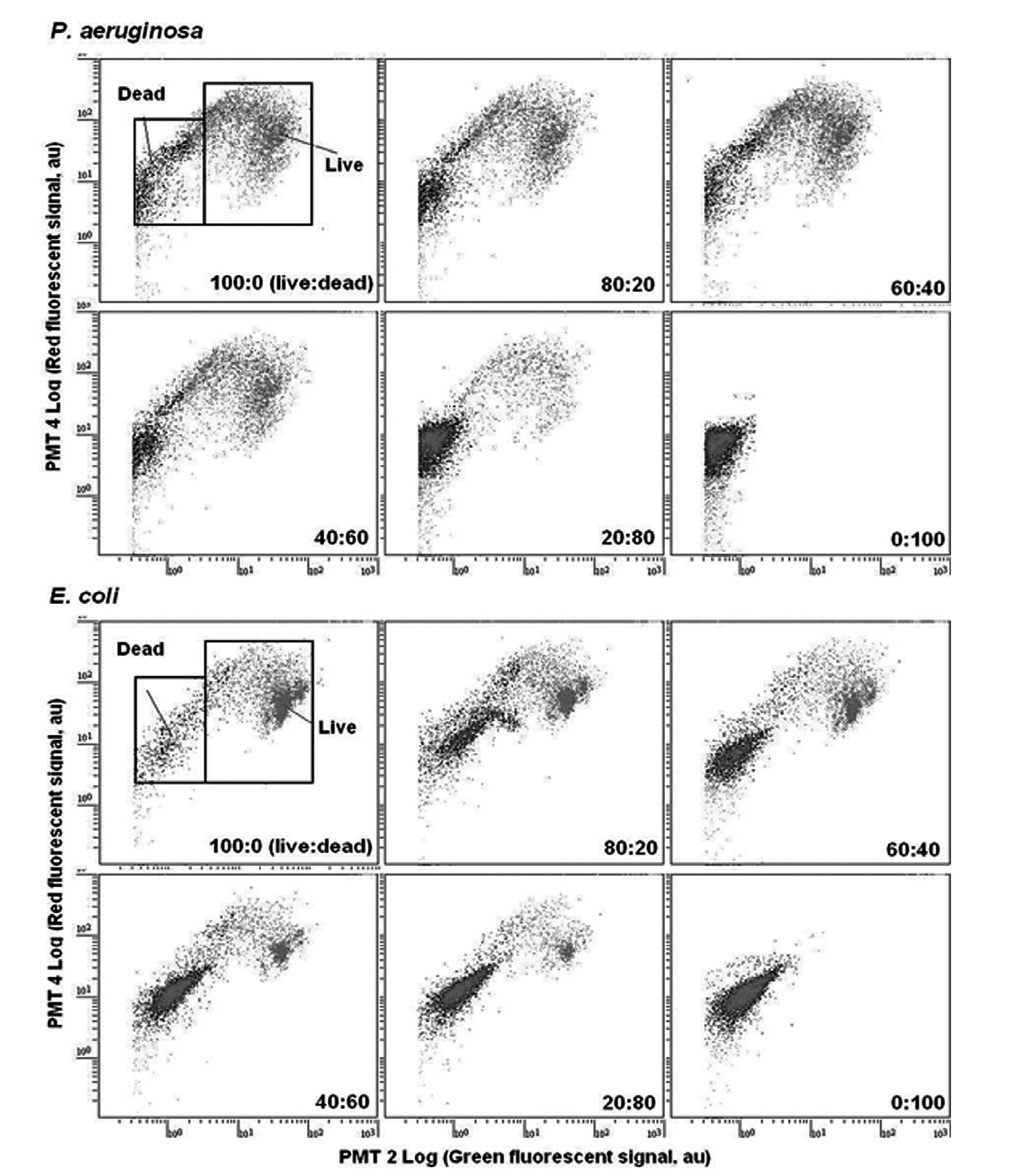
Fig. 1 showed overlays of fluorescence histograms of
The mixture of only SYTO 9 labeled model bacteria (live) and both SYTO 9 and PI labeled bacteria (dead) with percentages of 100:0, 80:20, 60:40, 40:60, 20:80, and 0:100 (live:dead) were then counted by multiparametric FCM performing region analysis on PMT 2 (green fluorescent, 510-540 nm) versus PMT 4 (red fluorescent, 615-640 nm) dot plots. As shown in the FCM cytograms
(Fig. 1), the gate for dead bacteria was defined as the range of 0.2 × 100 to 6.0 × 101 PMT 2 fluorescence (X-axis) and 2.0 × 100 to 2.0 × 102 PMT 4 fluorescence (Y-axis), and the gate for live bacteria was defined as the range of 6.0 × 100 to 6.0 × 102 PMT 2 fluorescence (X-axis) and 2.0 × 100 to 4.0 × 102 PMT 4 fluorescence (Y-axis). The number of live and dead cells could be calculated by creating gates on the cytograms. Consequently, using the FCM protocol with PMT 2 and PMT 4, live and dead
3.2. Application to the Monitoring of Bacterial Diagnosis
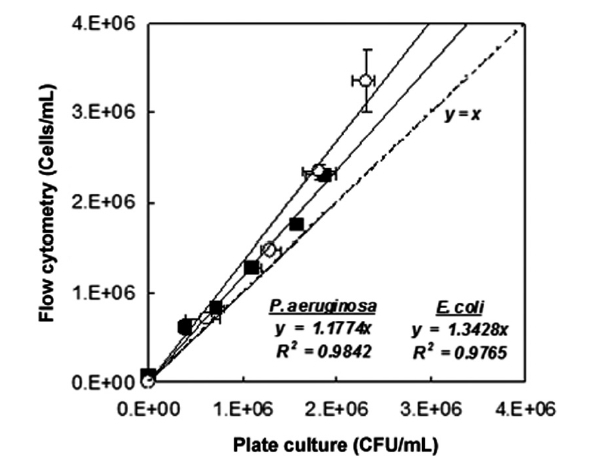
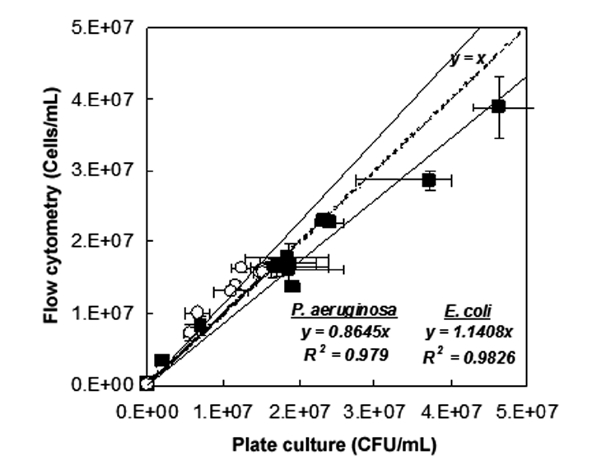
The results of fluorescence counts of bacterial diagnosis (viability) based on the region analysis on PMT 2 (green fluores-cent, 510-540 nm) versus PMT 4 (red fluorescent, 615-640 nm) dot plots were compared with the counts that were measured by the plate culture method (culturability assessment) as shown in Fig. 2. Additionally, the number of the tested bacteria in the live condition detected by FCM (cells/mL) well represented the number of those detected by culturability assessment with agar plate (CFU/mL). As the results on fitting the data with linear functions, slope was measured in 1.1774 cell/CFU (R2 = 0.9842, n = 6) in the case of
Consequently, the numbers of the tested bacteria detected by FCM protocol was slightly over-estimated than the number detected by the culturability assessment. These phenomena might be come from the physiological properties of bacteria that entered into a viable but non-culturable (VBNC) state due to the environmental or nutritional stresses. Hwang et al. [2] demonstrated that the culturability assessment based on the counting colony formed on agar plate was very difficult if the bacteria had entered into a state of VBNC. According to the observations, detection of viability as well as the density of the pure-cultured
Based on the observed results, FCM was surveyed to monitor the disinfection properties of
Compared with the data obtained from alcohol-fixed tests, the number of
In this study, multiparametric (PMT 2 for green fluorescence and PMT 4 for red fluorescence) FCM was installed to enumerate the bacterial diagnosis (viability as well as the population) of
The FCM protocol with PMT 2 and PMT 4 successfully determined diagnosis (live and dead) of
Consequently, it is concluded that application of FCM to monitor the functional effect of disinfectants on the physiological status of target bacteria can offer more rapid and reliable data compared to the plate culture colony counting method.