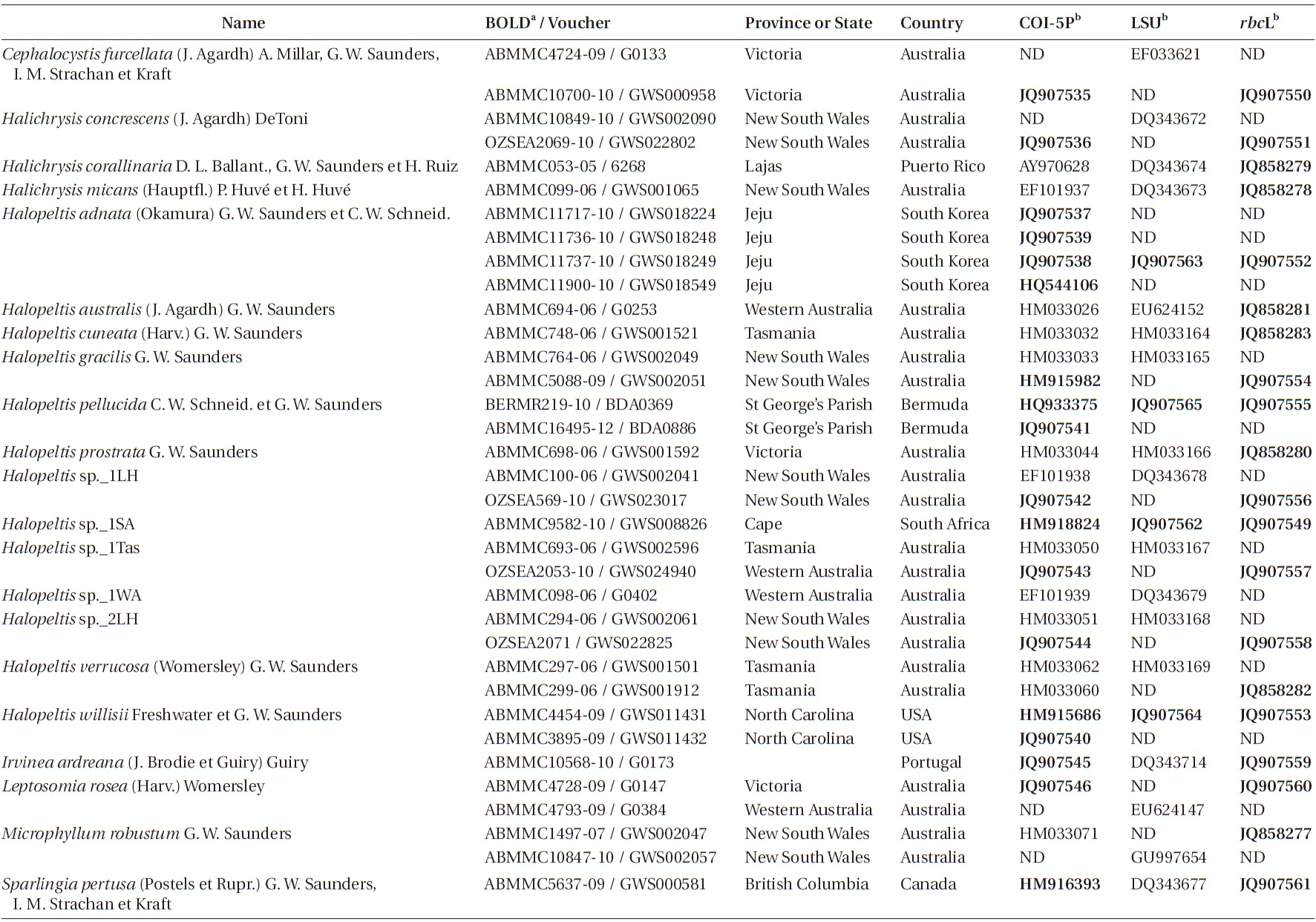
Significant attention has been paid of late to the Rhodymeniales using genetic sequencing to elucidate novel taxa and phylogenetic relationships among congeners. These studies have discovered cryptic diversity within the order (e.g., Saunders et al. 2006, Ballantine and Lozada- Troche 2008, Schneider and Lane 2008, Ballantine et al. 2010, Lozada-Troche and Ballantine 2010, Saunders and McDonald 2010), as well as allowing for generic discovery and realignment in the Rhodymeniaceae, Fryeellaceae, Faucheaceae, Hymenocladiaceae, Champiaceae and Lomentariaceae (e.g., Millar et al. 1996, Saunders et al. 1999, 2006, 2007, Wilkes et al. 2005, Dalen and Saunders 2007, Le Gall et al. 2008, Lozada-Troche et al. 2010, Saunders and McDonald 2010, Suzuki et al. 2010). Combined, such molecular-assisted, alpha taxonomic (MAAT) studies have begun to demonstrate a significantly clearer picture of species richness and evolutionary relationships within this order than was known in the 20th century (Bliding 1928, Kylin 1931, 1956, Sparling 1957).
Despite the many genera and species of red algae that have been investigated using MAAT over the past two decades, some taxa already sequenced require addi-tional collections and genetic material to clarify species relationships (e.g., Saunders and McDonald 2010, Kraft and Saunders 2011). Using mitochondrial barcoding techniques and large subunit ribosomal RNA (LSU) gene sequences, a recent paper by Saunders and McDonald (2010) resurrected the genus Halopeltis J. Agardh (1854), for the last 150 years an entity subsumed in Rhodymenia Greville (1830). Although Agardh (1854) properly validated this genus, within two decades he no longer used the generic name in his synoptic “Species genera et ordines algarum...” (Agardh 1876), by which time he reverted to recognizing Rhodymenia australis (Agardh 1876, p. 332). Although Agardh (1876) listed in synonymy the species Acropeltis australis J. Agardh (1852) for which he later established the genus Halopeltis (Agardh 1854), he did not mention his own Halopeltis there, or elsewhere in the section of Rhodymenia, the name apparently slipping into synonomy without notice. Subsequent 19th century phycologists made no mention of Halopeltis. The convoluted nomenclatural history of this genus is recounted in detail by Saunders and McDonald (2010), who, after resurrecting Halopeltis from synonomy, moved three additional Rhodymenia and Rhodymeniocolax Setch. (1923) species to the genus and described two novel species that they had collected in southeastern Australia and Lord Howe Island. All six known Halopeltis species have Southern Hemisphere distributions, with only the generitype, H. australis (J. Agardh) G. W. Saunders, broadly distributed throughout the Indo-Pacific, including reports without molecular confirmation in tropical waters of the Northern Hemisphere (Guiry and Guiry 2012).
DNA sequence analyses of recent collections in the Northern Hemisphere from warm temperate waters of Korea, Bermuda and North Carolina, USA showed them to phylogenetically group with Halopeltis. Morphological studies of these three species show that the Korean specimens matched the previously described Rhodymenia adnata Okamura from Japan and Korea, and that the two Atlantic entities represented novel species. In this paper we describe these entities using morphological and molecular approaches and characterize them in the context of the genus Halopeltis and the Rhodymeniaceae.
Collections were made by scuba, and specimens were dried fresh onto herbarium paper with portions dried in silica gel and others preserved in 4-5% Formalin-seawater. The site locations were taken above water using a Garmin GPS III Plus (Garmin, Olathe, KS, USA). Thin sections were made using a manual American Optical freezing microtome (American Optical, Buffalo, NY, USA) or by hand and these were stained and mounted in a 20 : 1 solution of 30% Karo corn syrup and 1% aniline blue with a drop of formalin added as a preservative. Field habit photographs were taken using a Nikon D50 digital camera (Nikon, Tokyo, Japan) or an Olympus 5060 digital camera (Olympus, Center Valley, PA, USA) with a Light & Motion housing (Light & Motion, Monterey, CA, USA) and Sea & Sea YS110 strobe (Sea & Sea, Long Beach, CA, USA). Photomicrographs were taken using Carl Zeiss Axioskop 40 microscope or Stemi 2000-CS (Carl Zeiss, Oberkochen, Germany) equipped with a model 4.2 Spot InSight QE digital camera (Diagnostic Instruments, Sterling Heights, MI, USA), a Zeiss Axio Imager.Z1 compound microscope fitted with an AxioCam MRc 5 camera system or an Olympus BX41 compound microscope fitted with a Roper Scientific Photometrics CoolSnap camera (Photometrics, Tucson, AZ, USA). The digital images were composed in Adobe Photoshop CS5 Extended version 12.0.2 (Adobe Systems, San Jose, CA, USA). Voucher specimens are variously deposited in KIRI, MICH, UNB, WNC and CWS’s personal herbarium. When listed, herbarium abbreviations follow the online Index Herbariorum (http://sweetgum.nybg.org/ih/) and standard author abbreviations follow Brummitt and Powell (1992).
Specimens used in molecular analyses are recorded in Table 1. After collection, specimens were silica-dried, and total DNA extraction followed Saunders and McDevit (in press) or the illustra Phytopure DNA Extraction Kit manufacturer’s recommended protocol (GE Healthcare, Buckinghamshire, UK). The COI-5P region of the mitochondrial genome (Saunders 2005, 2008) was first amplified to assign specimens to genetic species groups (Saunders and McDevit in press). The actual primer pair used to amplify this region for each sample is recorded with the sequence on GenBank (Table 1). For phylogenetic analyses, the partial plastid RUBISCO large subunit (rbcL) was amplified following Vis et al. (2007) with the primers F57 and rbcLrevNew (see Kucera and Saunders 2012), while the large subunit ribosomal DNA (LSU) was amplified with the primers and profile outlined in Harper and Saunders (2001). All PCR products were sequenced using a Big Dye Terminator Cycle Sequencing Ready Re
actions DNA sequencing kit (Applied Biosystems, Foster City, CA, USA). Final sequences (excluding the 5′ and 3′ primer regions) for all three markers were assembled using Sequencher TM 4.10 (Gene Codes Corp., Ann Arbor, MI, USA) and based on bidirectional data.
Multiple alignments were prepared in MacClade version 4.06 (Maddison and Maddison 2003). For species identification, a neighbor joining analysis of the COI-5P alignment with uncorrected distances was completed using PAUP* (Swofford 2003) in Geneious (Drummond et al. 2009). For phylogenetic analyses the individual gene regions (COI-5P, rbcL, and LSU), as well as a combined multigene alignment, were subjected to maximum likelihood (ML) analysis with a GTR + I + G model of evolution using PHYML in Geneious (Drummond et al. 2009). Robustness was assessed through analyses of 500 bootstrap repetitions. For the multigene alignment MrBayes (v. 3.1.2) (Huelsenbeck and Ronquist 2001) was additionally used to complete two independent trials (each with parallel runs) of Bayesian inference under a GTR + I + G model. Parallel runs of four Markov chains were completed with one million generations and sampling each 100 generations. Data were partitioned by gene, and then by codon for rbcL and COI-5P, and the parameters were unlinked with the overall rate allowed to vary across partitions. The burn-in for each run was determined by plotting overall likelihood scores against generation, which established the stationary phase of each run for estimating the posterior probability distribution - the final estimate based on pooled samples from two independent runs. We were unable to obtain rbcL data for Halopeltis sp._1WA and thus we completed both the ML and bayesian multigene analyses in duplicate: with these data coded as missing and with this species removed from the alignment. This allowed us to assess the impact of the missing data on the overall topology and support.
Collections of unknown rhodymeniaceous taxa (Table 1) from Bermuda (n = 2; Halopeltis pellucida), North Carolina (n = 2; H. willisii) and Korea (n = 4; H. adnata) were subjected to routine DNA barcode screening and formed three distinct genetic species groups. For all three genetic groups, the barcode marker was identical (0% divergence) for all individuals within a species, whereas divergence between these three genetic groups ranged from 7.8 (between H. adnata and H. willisii) to 16.1% (previous two to H. pellucida). Finally, blast searches at BOLD and Genbank both indicated an alliance of these three novel genetic groups with species of the genus Halopeltis (data not shown).
To explore further the phylogenetic affinities of the newly discovered COI-5P genetic species, LSU and rbcL data were generated for one representative each of these three newly uncovered genetic species, as well as for additional rhodymeniaceous species (Table 1); emphasizing the Halichrysis and Halopeltis lineage with suitable outgroups (Saunders and McDonald 2010), to allow for single (COI-5P, LSU, and rbcL), as well as a combined gene analyses (Fig. 1). Within the limits of statistical support, all single gene analyses resulted in trees similar to that obtained in a combined dataset analysis, although levels of support were typically lower (not shown). Further, missing rbcL data for Halopeltis sp._1WA did not significantly change the resulting tree (topology or support levels). Thus, we present only the phylogenetic results of the multigene (including Halopeltis sp._1WA with rbcL data recorded as missing) analyses for ML with bootstrap support and posterior probabilities from the Bayesian analyses appended (Fig. 1). Our analyses solidly resolved the genera Halichrysis and Halopeltis as reciprocally monophyletic sister taxa consistent with published studies. Our three novel genetic species groupings resolved with the latter genus, two of them, H. adnata and H. willisii, forming an alliance with the deeply diverging Halopeltis sp._1WA, while the third, H. pellucida, resolved strongly with a collection from South Africa (Table 1), Halopeltis sp._1SA (Fig. 1). The last mentioned species was tentatively field-identified as Botryocladia madagascariensis G. Feldmann and likely represents the first report of a Halopeltis with inflated axes. This species requires further taxonomic study.
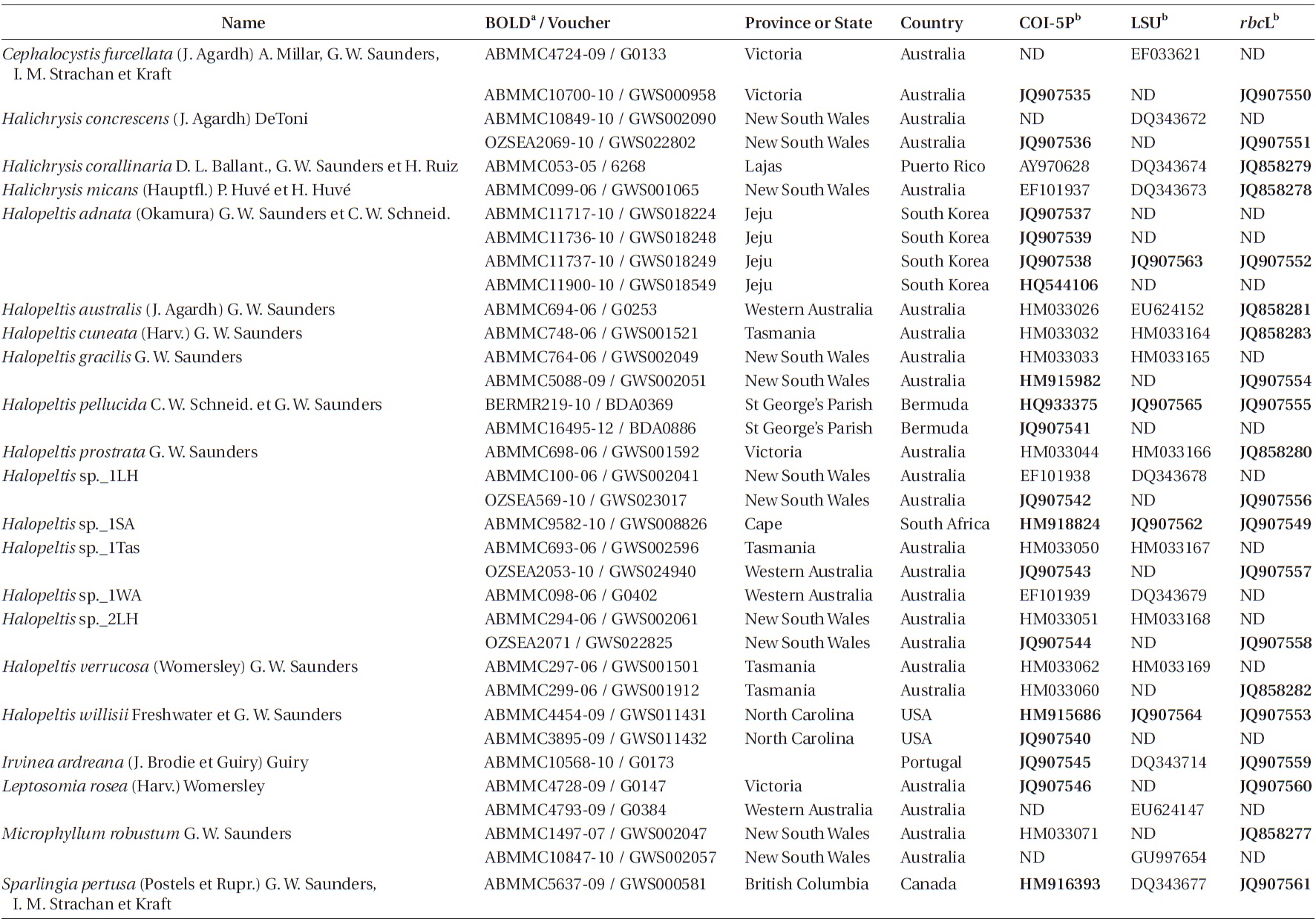
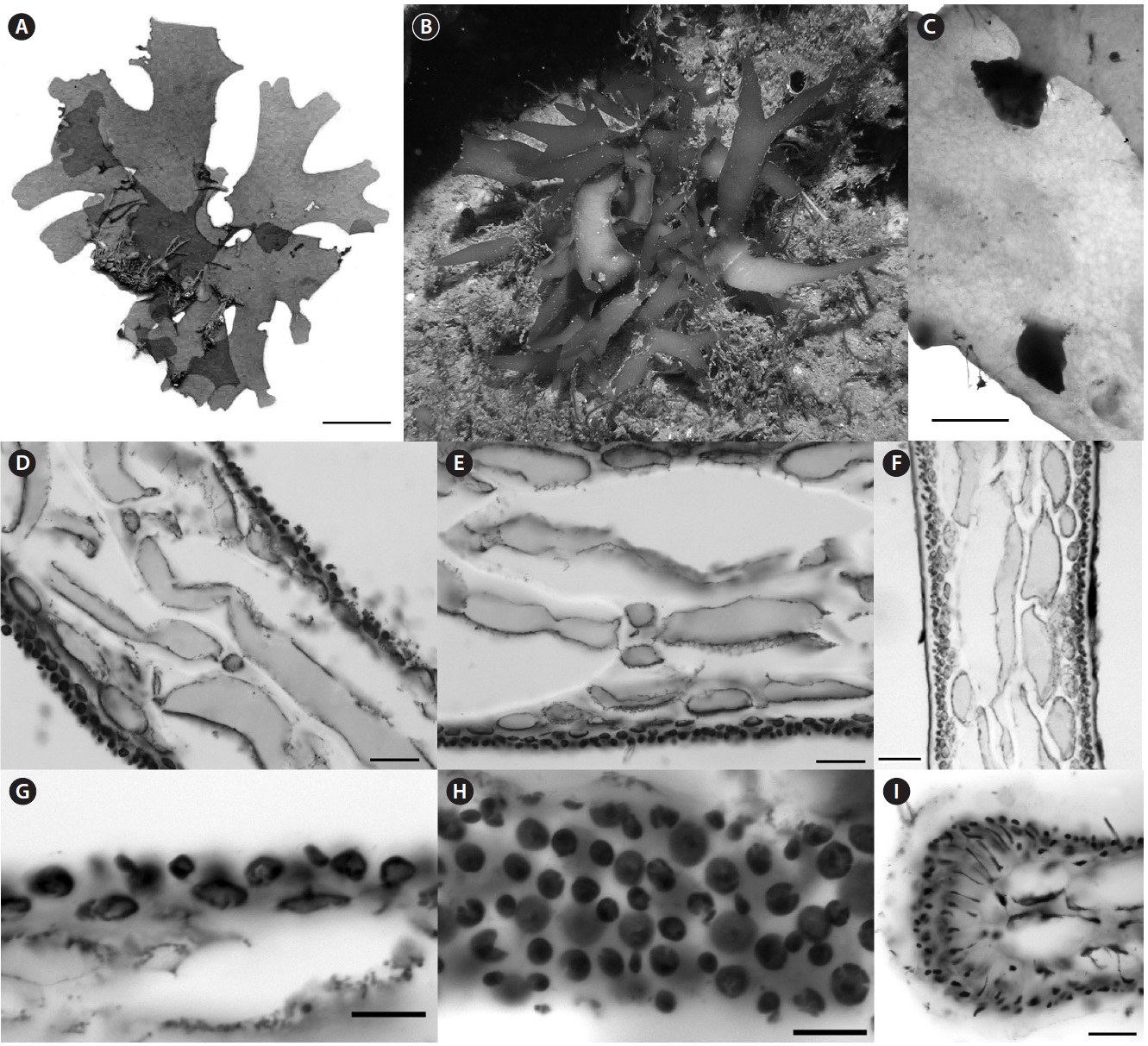
Recent collections of Rhodymenia adnata Okamura from 10-15 m off Jeju Island, Korea (Table 1) closely match the protologue (Okamura 1934) and other published descriptions for this species (e.g., Lee 2008), as well as general features recently illuminated for the genus Halopeltis (Saunders and McDonald 2010). Plants range from 2.5-8.0 cm in diameter and are flat, membranous, procumbent and attached to the substratum and adnate to each other by attachment pads (Fig. 2A). Although slightly smaller than reported for the type (11 cm measured from Okamura 1934, Pl. 319, Fig. 1), the Korean collections match
R. adnata in all of the other gross morphological features. The color of fresh plants range from reddish in parts to purplish red variegated with tints of green and expanses of yellow. The blades are strongly iridescent when fresh, agreeing with the protologue in all but the last attribute (Okamura 1934). In this regard, our Korean collections differ from those described in Lee (2008, p. 405), which are simply described as red (possibly an artifact of fixation), but which nonetheless share all of the other gross morphological features outlined here.
Anatomically, blades of the Korean material are 200-300 μm in width near the margins and composed of a medulla of 2-3 axially elongate medullary cells, a 1-2 layered inner cortex of refractive cells and an outer cortex of rounded to slightly elongate outer cortical cells as documented in Lee (2008, p. 405, Fig. D). In middle portions of the thallus, the blades thicken to 300-325 μm, but are composed of essentially the same number of cell layers. In these sections, however, spaces invested with chains of smaller intercalating cells were now evident between the large medullary cells (Fig. 2B). This feature is consistent with species of the genus Halopeltis and, although not discussed in the protologue of Rhodymenia adnata, is nevertheless diagrammed by Okamura (1934, Pl. 319, Fig. 3) and documented by Lee (2008, p. 405, Fig. C) in reporting this species from Korea. As illustrated in the protologue (Okamura 1934, Pl. 319, Fig. 6), the outer cortex is incomplete with the underlying inner cortical cells clearly visible in surface view.
Tetrasporangia were unknown at the time of the original description of Rhodymenia adnata, but were later reported by Lee (2008, p. 405, Fig. C). One of the recent Korean collections is tetrasporangial. Our new observations are consistent with those of Lee (2008), and are consistent with species included in the genus Halopeltis (Saunders and McDonald 2010). Tetrasporangial sori are relatively extensive patches at the branch apices that follow the thallus outline (as in Halopeltis; Fig. 2A) and are formed on a single thallus surface [upper according to Lee (2008); see Saunders and McDonald (2010, p. 657) for a description of sorus placement in other Halopeltis spp.]. In Rhodymenia, tetrasporangia derive from inter
calary cortical cells at the transition between inner and outer cortical layers (Saunders et al. 1999, Saunders and McDonald 2010). Sparling (1957, Fig. 12) and Guiry (1974, Fig. 1) illustrate this feature for the generitype, R. pseudopalmata (J. V. Lamour.) P. C. Silva, and other species currently recognized in the genus. The tetrasporangia in the Korean plants derive from the direct conversion of outer cortical cells without adventitious growth or paraphyseal development and with sporangial initials commonly and clearly pit-connected to inner cortical cells (Fig. 2C). Again, although not reported by Lee for R. adnata, this mode of tetrasporangial development is consistent with his documented images (Lee 2008, p. 405, Fig. C) and the genus Halopeltis (Saunders and McDonald 2010). Mature tetrasporangia were cruciately divided and ranged in size from 15-20 μm in diameter and 22-28 μm long (Fig. 2D), once again consistent with the observations in Lee (2008) and with the genus Halopeltis (Saunders and McDonald 2010).
Taking all of this in consideration, our recent Korean collections of Rhodymenia adnata clearly bring to mind the genus Halopeltis, and this is consistent with their clustering with this genus in our molecular trees (Fig. 1). However, R. adnata and its allies (vis., Halopeltis sp._1WA and H. willisii) resolved as sister to, rather than embedded among, the included species of Halopeltis in our phylogenies, which leaves open the possibility that these species actually belong to a sister genus of Halopeltis. Given their many shared morphological features, however, we consider the conservative taxonomic decision at this time is the transfer of R. adnata to Halopeltis, which is implemented here:
Basionym. Rhodymenia adnata Okamura 1934, p. 35-36, Pl. 319.
Interestingly, this reassignment to Halopeltis in effect represents the first species in the genus from non-tropical waters of the Northern Hemisphere. The only member of the Southern Hemisphere genus distributed north of the equator is the type, H. australis, which has been reported from tropical regions of the Indo-Pacific (Silva et al. 1996, as Rhodymenia sonderi P. C. Silva; Guiry and Guiry 2012), although such records await molecular confirmation.
Other recent western Atlantic Ocean collections of rhodymeniacean specimens from North Carolina, USA (34° N) and Bermuda (32° N) were also shown to group in a clade with Halopeltis in our COI-5P, LSU and rbcL consensus phylogenetic tree (Fig. 1). As neither of these morphotypes could be accommodated by any of the known species of Halopeltis, we herein establish new species for them, each representing, along with H. adnata, species from the Northern Hemisphere.
Description. Blades procumbent, flattened, anastomosing and pseudodichotomously branched, to 80 mm long and to 15 mm wide near the base, narrowing to 4-5 mm wide in ultimate branches, rosy-brown to pink in situ; attached by haptera scattered along margins and ventral surface, margins otherwise smooth; cortex composed of 2-3 layers of pigmented cells, outer cortex incomplete, cells globose to quadrangular, 4-10 μm in diameter, inner cortical cells transversely elliptical, 10-17 μm in greatest dimension; central medulla composed of 2-3 layers of lightly-pigmented, thin-walled, highly vacuolate, subglobose to polygonal, somewhat axially elongated cells up to 350 μm in diameter, with interspersed, variably shaped, smaller 10-200 μm diameter cells at some interstices; outer medullary cells transversely elliptical, 40-100 μm in diameter.
Tetrasporangia cruciately divided, 15-25 μm in diameter and 30-40 μm long, developmentally replacing outer cortical cells and pit-connected to inner cortical cells in variably shaped sori on one or both blade surfaces, surrounded by anticlinally elongated outer cortical cells that sometimes form fasciculate clusters.
Cystocarps developing on both blade surfaces and margins, hemispherical, not basally constricted, somewhat flattened at the ostiole; pericarp 8-15 cells thick, locule wall lined with network of rounded cells, and network of darkly staining nutritive cells that develop in the chamber floor; initial gonimoblast cells large, darkly staining, giving rise to filamentous gonimoblasts that ramify among closely adjacent lobes of densely packed obovate to angular carposporangia, 12-20 μm in diameter.
Etymology. The epithet honors James Newman Willis III, “Capt. Jim,” US Bureau of Commercial Fisheries and National Marine Fisheries researcher, coastal advocate and historian scholar of North Carolina’s Outer Banks.
Holotype (deposited in UNB). NCweed-178, ?, 30 June 2009, Southwest Ledge, Onslow Bay, North Carolina, USA, western Atlantic Ocean, 34°23.576′ N, 076°53.924′ W, on rock, depth 27 m, coll. A. Poray [Isotype, WNC31611].
Paratypes. NCweed-819, ?, 01 August 2007, Witzig Rock (“West Rock”), Onslow Bay, NC, 34°16.668′ N, 076°34.949′ W, on rock, depth 30 m, coll. D. W. Freshwater & B. Deagan [WNC32155]; NCweed-820, female, 01 August 2007, Witzig Rock, loc. cit., coll. D. W. Freshwater & B. Deagan [WNC32154].
Plants of Halopeltis willisii are shortly procumbent with few to many pseudodichotomously branched blades that are sometimes basally anastomosing (Fig. 3A & B). Blade bases are attached by multiple peg-like, secondary holdfasts that develop from the margins and ventral surfaces (Fig. 3C). Occasionally, distal portions of blades also develop secondary attachments to the substrate. The blades are constructed of two or three layers of large, subglobose or polygonal to transversely elliptical inner medullary cells with interspersed smaller cells within many interstices; an outer medulla of smaller transversely elliptical cells, and a surrounding, more heavily pigmented cortex (Fig. 3D-F). Inner cortical cells are circular in surface view but transversely elliptical and give rise to an incomplete outer cortex of globose to quadrangular cells (Fig. 3G & H). Inner cortical cells are, on occasion, anticlinally elongated at the blade margins (Fig. 3I), perhaps where attachments are developing, as sections through holdfasts reveal similarly elongate cells.
Tetrasporangial sori are variously shaped and develop on both blade surfaces (Fig. 4A & B). They are not strongly nemathecial in appearance as described for Halopeltis australis (J. Agardh) G. W. Saunders (as Rhodymenia australis Sond.) and H. verrucosa (Womersley) G. W. Saunders (as Rhodymenia verrucosa Womersley; Womersley 1996). Tetrasporangia are produced by the conversion of outer
[Fig. 3.] Halopeltis willisii sp. nov. (A) Holotype specimen, NCweed-178. (B) In situ image of specimen NCweed-819. (C) Peg-like holdfasts on ventral surface and margin of blade, NCweed-819. (D) Longitudinal section showing large inner medullary cells with smaller cells in some interstices and the outer medulla, NCweed-819. (E) Longitudinal section showing large inner medullary cells with smaller cells in some interstices and the outer medulla. Membranes of large cells were disrupted during sectioning causing the cytoplasm to look plasmolyzed, NCweed-819. (F) Transverse section showing large inner and smaller outer medullary cells, NCweed-820. (G) Transverse section showing transversely elliptical inner cortical cells giving rise to multiple outer cortical cells, NCweed-819. (H) Blade surface view with more darkly stained outer cortical cells (upper focal plane) forming an incomplete outer cortex over the inner cortical cells (lower focal plane), NCweed-819. (I) Transverse section through blade margin where inner cortical cells are anticlinally elongate, NCweed-178. Scale bars represent: A, 1 cm; C, 0.25 mm; D-F & I, 50 μm; G & H, 20 μm.
cortical cells (Fig. 4C). Non-fertile outer cortical cells become elongate and form fasciculate clusters borne on the inner cortical cells (Fig. 4C).
Neither pre-fertilization nor immediate post-fertilization carpogonial and auxiliary cell branches were observed. Cystocarps develop on both blade surfaces and margins, and are hemispherical with no basal constriction (Fig. 4D-F), the latter trait considered uncommon in the genus (Saunders and McDonald 2010). The pericarp and locule begin forming early in the post-fertilization process and a basal nutritive tissue of darkly staining cells develops in the floor of the expanding cystocarp (Fig. 4E & G). The auxiliary cell cuts off a gonimoblast initial (Fig. 4H & I), which will produce gonimoblast filaments
[Fig. 4.] Halopeltis willisii sp. nov. (A) Tetrasporangial sori on blade surface, NCweed-819. (B) Transverse section with tetrasporangial sori on both blade surfaces, NCweed-178. (C) Longitudinal section through a tetrasporangial sorus with fasciculate clusters of outer cortical cells and a developing tetrasporangia pit-connected (arrow) to an inner cortical cell, NCweed-819. (D) Section through a young cystocarp with the gonimoblast initial (arrow) visible among the developing basal nutritive tissue (more darkly staining cells) and at the base of the developing ostiole, NCweed-820. (E) Section through a developing cystocarp with darkly staining network of basal nutritive cells and remnant connections between inner pericarp cells (arrows) visible, NCweed-820. The carposporophyte was dislodged during sectioning. (F) Section through a mature cystocarp with a thick pericarp, darkly staining gonimoblast cells and tightly packed carposporangia, NCweed-820. (G) Section through a mature cystocarp showing the flattened darkly staining nutritive tissue cells, filamentous gonimoblast cells (arrows) within tightly packed carposporangia and remnant connections between inner pericarp cells (arrowheads), NCweed-820. (H) Section of early post-fertilization cystocarp showing an auxiliary mother cell (amz) and auxiliary cell (az), NCweed-179. (I) Section of an early post-fertilization cystocarp with a gonimoblast initial (gi) and auxiliary cell (az), NCweed-179. (J) Two carposporangia pit-connected (arrow) to a darkly stained filamentous gonimoblast cell, NCweed-820. Scale bars represent: A, 1 mm; B, 100 μm; C & H-J, 20 μm; D, E & G, 50 μm; F, 100 μm.
that are darkly staining and ramify among lobes of densely packed carposporangia (Fig. 4F, G & J). The inner pericarp cells form an interconnected network but remain rounded as has been described for Halopeltis prostrata G. W. Saunders (Saunders and McDonald 2010). Remnant stretched connections are persistent between the upper and lower inner pericarp cells (Fig. 4E & G). Cells of the basal nutritive tissue become flattened as cystocarps mature (Fig. 4F & G).
The habit of Halopeltis willisii is much broader than all of the known species of Halopeltis, and in fact could be superficially confused with young specimens of Agardhinula browneae (J. Agardh) De Toni, a species likewise known from deep offshore waters of North Carolina (Schneider and Searles 1991). Despite not having material of the much larger, erect A. browneae available for genetic sequencing at present, it does have morphological and reproductive characteristics distinct from H. willisii. Although the medulla of A. browneae has variously sized cells (Schneider and Searles 1991), they do not develop small cells at the interstices of adjacent cells as in H. willisii (Fig. 3D & E). Using preserved material of A. browneae in the first author’s herbarium, we have observed tetrasporangia developing directly from surface cortical cells in massive numbers of sori, the sori often becoming confluent and appearing as a reticulated pattern covering all but the most apical regions of the blades. These sori are more darkly pigmented than the vegetative outer cortex, and give the blades a distinctly mottled appearance when fully developed (Schneider and Searles 1991). In H. willisii, tetrasporangia are formed in smaller, usually discrete, sori, and are surrounded by anticlinally elongated outer cortical cells. In A. browneae, paraphyses form from outer cortical cells to surround the sporangia in elevated gelatinous nemathecia. The ontogeny of tetrasporangia and nemathecia in A. browneae was observed by Saunders and McDonald (2010), and they suggested it was similar to that outlined for the Fryeellaceae (Le Gall et al. 2008) rather than the Rhodymeniaceae where it was placed in the past (Schneider and Wynne 2007). Without knowledge of tetrasporangial development in Agardhinula, Le Gall et al. (2008) had considered the genus of uncertain position within the Rhodymeniales.
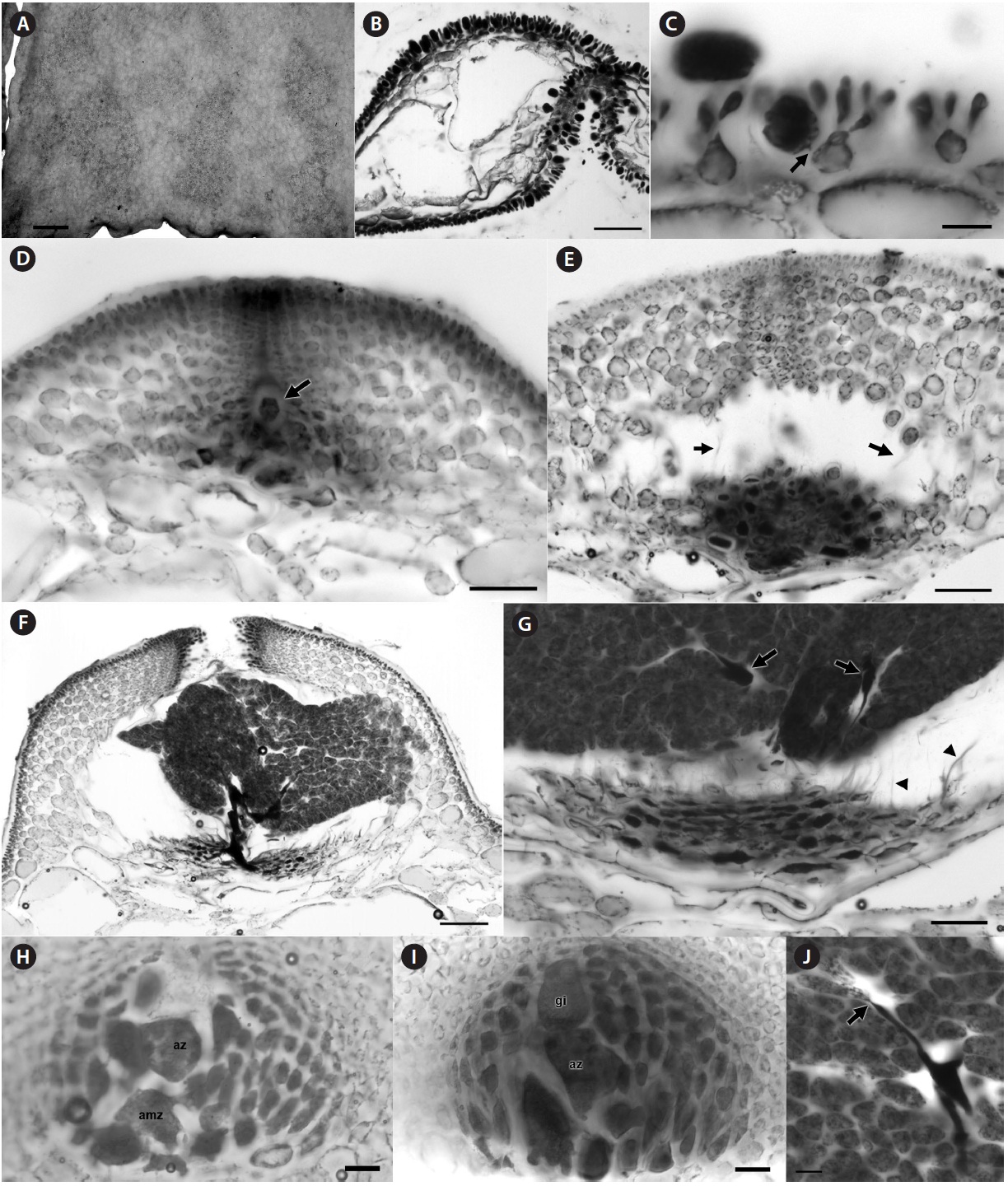
Description. Plants procumbent and translucent, spreading to 2 cm across; blades ligulate, arising from short branched stipes, pseudodichotomously branched; blades 2.0-2.5 mm in diameter in ligulate portions and to 4.0 mm in diameter below branches, to 260 μm thick; attached initially to substratum by a small basal holdfast, and secondarily to the substratum and themselves by haptera issuing from blade margins, margins otherwise smooth; blade apices rounded to spatulate (Fig. 5A & B); cortex incompletely forming over the interstices of outer medullary cells in surface view in apical regions (Fig. 5E), composed of 1-2 layers of circular to ellipsoidal pigmented cells from 5-12 μm in greatest dimension (Fig. 5F); cortex connecting to a lightly-pigmented pseudoparenchymatous medulla with 2-3 layers of larger globose to rounded polygonal cells (Fig. 5F&I) with strongly reticulated cytoplasts (Fig. 5E & F), cells from 25-120 μm in greatest dimension.
Tetrasporangia cruciately divided, broadly ellipsoidal, 12-22 μm in diameter and 17-31 μm long (Fig. 5G & H), developmentally replacing outer cortical cells and pit-connected to inner cortical cells in oval to cordiform, raised nemathecial-like sori forming at the apices of blades or below branch forkings (Fig. 5B), surrounded by anticlinally elongated outer cortical cells (Fig. 5G & H); sori forming on the upper surface.
Gametophytes monoecious; spermatangia and cystocarps developing on both blade surfaces; cystocarps hemispherical, somewhat constricted basally, ostiolate, sometimes grouped contiguously; pericarp 8-12 cells thick, ostiole wall lined with a network of densely pigmented, flattened cells; carposporophyte compressing medullary cells below the cystocarp, forming a network of darkly staining nutritive cells that develop in the chamber floor (Fig. 5M); gonimoblast cells small, darkly staining, giving rise to secondarily pit-connected gonimoblast cells that ramify and produce obovoid carposporangia to 12 μm in diameter; spermatangia in sori at times ringing the cystocarps (Fig. 5I), even slightly covering the basal portions of the pericarp (Fig. 5J); spermatangia ellipsoidal, 2-3 μm in diameter (Fig. 5K & L).
Etymology. The epithet pellucida (L, fem.), represents the new species’ translucent, but not hyaline, ligulate blades.
Holotype (deposited in UNB). CWS10-24-6 [DNA0369], ?, 23 August 2010, at Cathedral Rock, off south shore of Bermuda Is., Bermuda, western Atlantic Ocean, 32°20′31.1″ N, 64°39′24.2″ W, depth 17m, coll. C. W. Schneider [Isotypes: MICH, Herb. CWS].
Paratype. TRP12-36-6 [DNA0886], monoecious gametophytes, 18 February 2012, Cathedral Rock, Bermuda, loc. cit., coll. T. R. Popolizio [KIRI, Herb. CWS].
The small prostrate plants representing Halopeltis pel
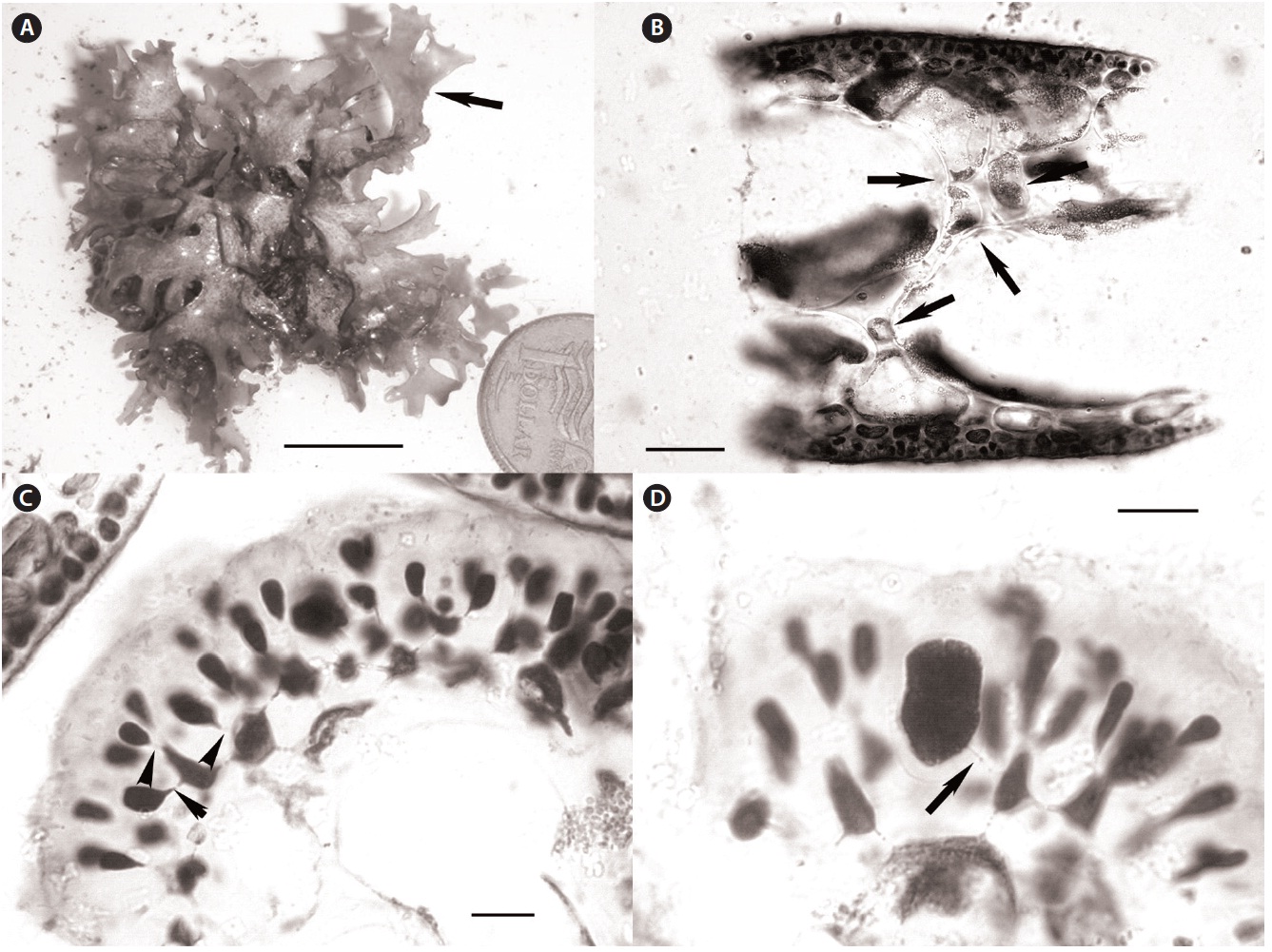
[Fig. 5.] Halopeltis pellucida sp. nov. (A & B) Holotype with tetrasporangial sori at apices and dichotomies (arrows), CWS10-24-6 [DNA0369]. (C) Habit, TRP12-36-6 [DNA0886]. (D) Habit of an isotype, CWS10-24-6. (E) Surface view of incomplete cortex showing medullary cells with strongly reticulated cytoplasts (arrows), CWS10-24-6. (F) Section at margin demonstrating spherical medullary cells and 1-2-layered cortex, CWS10-24-6. (G & H) Sections through a sorus showing tetrasporangia pit-connected to inner cortical cells (arrows) and separated by anticlinally elongated outer cortical cells, CWS10-24-6. (I) Cross section of a mature cystocarp ringed by a spermatangial sorus (arrows), TRP12-36-6. (J) Section through a pericarp wall showing basal constriction and a spermatangial sorus ascending up the cystocarp (arrows), TRP12-36-6. (K) Slightly raised spermatangial sorus in section, TRP12-36-6. (L) Spermatangial sorus in surface view, TRP12-36-6. (M) Flattened medullary cell below the carposporophyte cutting off nutritive cells (arrows). Scale bars represent: A, C & D, 5 mm; B, 100 μm; E & F, 50 μm; G, H & K-M, 25 μm; I & J, 200 μm.
lucida (Fig. 5A, C & D) were discovered offshore on heavily shaded horizontal rocks during late summer and middle winter in Bermuda. These delicate specimens were so translucent underwater that they were not discovered in the first collection until back in the lab where they were processed. The same site was revisited twice in winter 2012, and one of these trips lead to a second collection of H. pellucida. As of now, sporophytes have been found in the summer and gametophytes in the winter.
One of the substantive characteristics outlined for Halopeltis by Saunders and McDonald (2010) to differentiate it from Rhodymenia and Pseudohalopeltis G. W. Saunders were small medullary cells filling some of the spaces between medullary cells in at least older portions of the blades of H. australis and the other species joining it in the resurrected genus. In some species, such smaller medullary cells budded off the larger cells are rare and not seen throughout, so this feature is variable among and between species (Saunders and McDonald 2010). The Cathedral Rock specimens do not demonstrate this feature (Fig. 5E), thus at this time H. pellucida is a representative of the genus not demonstrating a feature common to its congeneric relatives. In older portions of H. pellucida blades, cross sections demonstrate a tightly packed medulla with the larger cells grading to smaller outer cortical cells, in all areas lacking substantive gaps between cells (Fig. 5F & I). It remains possible that the intercalating cells in other Halopeltis species only form because of the available spaces between large medullary cells, spaces not obvious in H. pellucida. H. pellucida produces medullary cells containing reticulate cytoplasts (Fig. 5E & F) reminiscent of many rhodymenialean species including two from Bermuda, Asteromenia bermudensis and A. peltata (Saunders et al. 2006), but until now not reported for Halopeltis.
As was the case for H. willisii, neither pre-fertilization nor post-fertilization carpogonial and auxiliary cell branches were observed for H. pellucida. In this species, cystocarps form on both blade surfaces and are often grouped in twos or threes. Cystocarps have slight basal constriction (Fig. 5I & J), a trait more typical of the genus (Saunders and McDonald 2010). The carposporophyte develops over a basal bed of darkly staining cells in the floor of the expanding cystocarp, and medullary cells below becoming flattened as carposporophytes mature (Fig. 5M). Gonimoblast cells elongate and pit-connect with other branches in the carposporophyte (Fig. 5M) before developing carposporangia. Inner pericarp cells become stretched as the pericarp enlarges, but remain connected to the growing carposporophyte (Fig. 5I).
H. pellucida is most similar in habit and size to H. gracilis G. W. Saunders a species from Lord Howe Is. This procumbent Pacific species has narrow branches (1.5-3.0 mm broad) from 110-190 μm thick (Saunders and McDonald 2010), narrower and thinner than those of H. pellucida (2-4 mm broad, to 240 μm thick). The tightly packed globose medullary cells of H. pellucida (25-120 μm in greatest dimension) are differently shaped than the ellipsoidal cells of H. gracilis (37-45 μm in diameter, 60-125 μm long), cells that unlike those in the new species are separated from each other by spaces filled by intercalating smaller cells (Saunders and McDonald 2010). Halopeltis pellucida along with H. willisii represent new additions to the marine flora of the Northern Hemisphere as well as the first members of the genus in the Atlantic Ocean, both now members of the western Atlantic warm temperate biogeographic region (Luning 1990).




![Halopeltis pellucida sp. nov. (A & B) Holotype with tetrasporangial sori at apices and dichotomies (arrows), CWS10-24-6 [DNA0369]. (C) Habit, TRP12-36-6 [DNA0886]. (D) Habit of an isotype, CWS10-24-6. (E) Surface view of incomplete cortex showing medullary cells with strongly reticulated cytoplasts (arrows), CWS10-24-6. (F) Section at margin demonstrating spherical medullary cells and 1-2-layered cortex, CWS10-24-6. (G & H) Sections through a sorus showing tetrasporangia pit-connected to inner cortical cells (arrows) and separated by anticlinally elongated outer cortical cells, CWS10-24-6. (I) Cross section of a mature cystocarp ringed by a spermatangial sorus (arrows), TRP12-36-6. (J) Section through a pericarp wall showing basal constriction and a spermatangial sorus ascending up the cystocarp (arrows), TRP12-36-6. (K) Slightly raised spermatangial sorus in section, TRP12-36-6. (L) Spermatangial sorus in surface view, TRP12-36-6. (M) Flattened medullary cell below the carposporophyte cutting off nutritive cells (arrows). Scale bars represent: A, C & D, 5 mm; B, 100 μm; E & F, 50 μm; G, H & K-M, 25 μm; I & J, 200 μm.](http://oak.go.kr/repository/journal/11244/JORHBK_2012_v27n2_95_f005.jpg)