Agar is a complex polysaccharide composed of agarose and agaropectin that originates from marine red algae such as
Agarolytic enzymes can be divided into two groups depending on their mode of action, α- and β-agarases, which produce agaro-oligosaccharides and neoagaro-oligosaccharides, respectively (Suzuki et al., 2002). Neoagaro-oligosaccharides possess various biological activities, including antitumor activity, macrophage-stimulating activity to increase immune function, anti-oxidizing activity, probiotic activity, and a skin moisturizing and whitening effect (Rhee et al., 2010). The agaro-oligosaccharide produced by α-agarase possesses anti-oxidative and anti-cancer activities (Ohta et al., 2005).
Some researchers have investigated the possibility of obtaining agarolytic enzymes from bacteria (Ohta et al., 2005; Lee et al., 2007). Although many genera of agarolytic bacteria have been identified, mainly from marine environments, since the first isolation from seawater by Gran in 1902, novel and more active agarolytic enzymes are required for industrial applications (Rhee et al., 2010). Therefore, to investigate the possibility of valuable industrial applications, we isolated a novel agar-degrading bacterium from seawater and characterized its culture properties and enzymatic activity.
>
Isolation of bacteria, media and chemicals
Seawater, seaweeds, and mud sampled from the seashore in Busan, Korea, were used for bacterial isolation. Media for isolation were marine broth 2216 and nutrient broth, which was prepared with 0.4 μm-filtered seawater and 1.5% agar. One hundred microliters of each sample was spread on an agar plate and incubated at 37℃ for five days. Lugol’s iodine solution (2 g KI and 1 g I2 in 200 mL water) was then sprayed onto the agar medium. Bacterial colonies that formed a clear zone were selected as agarolytic bacteria. Selected colonies were stored at -80℃ with glycerol until required. Media used in this study were obtained from Difco Co. (Detroit, MI, USA) and chemicals were purchased from Sigma Co. (St. Louis, MO, USA).
>
Identification of isolated strain
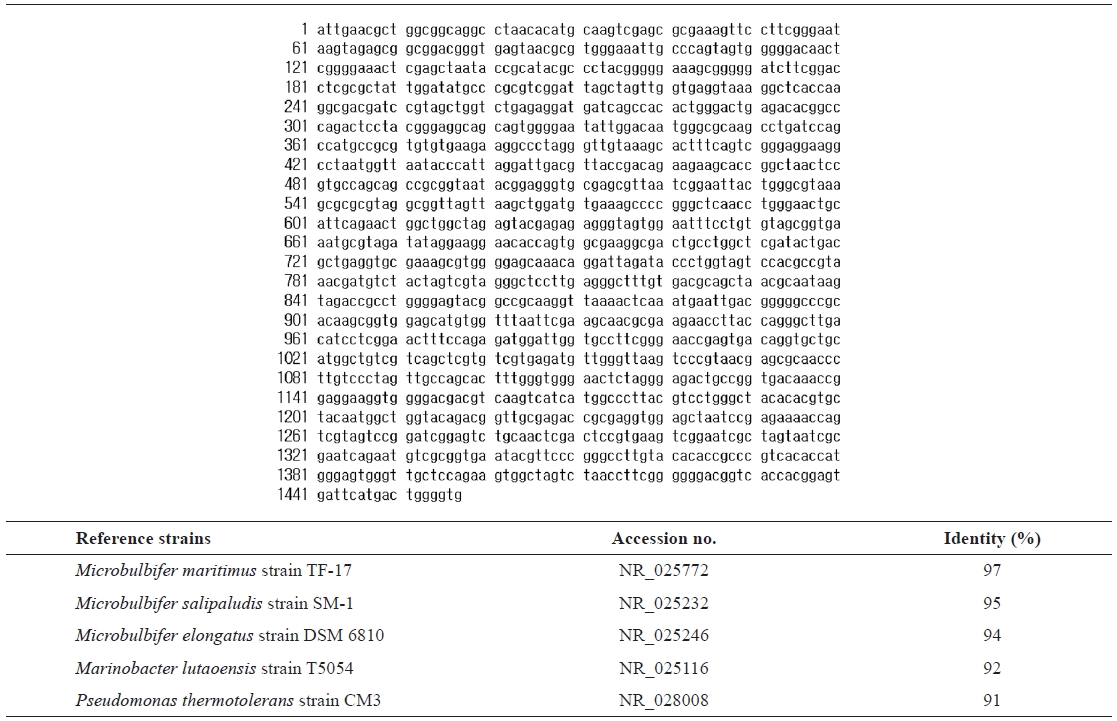
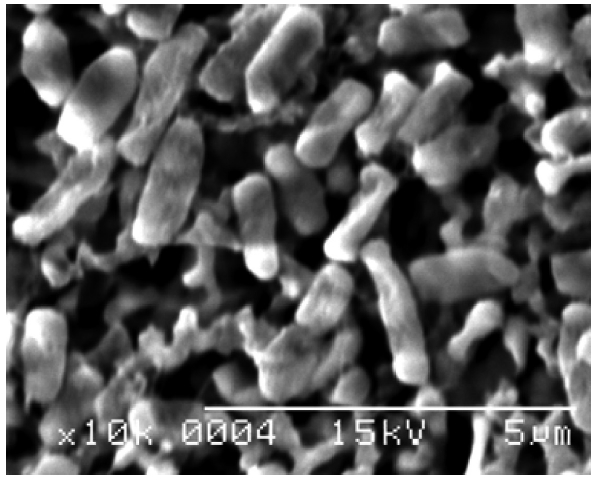
The isolate was identified by 16S rRNA sequencing and scanning electron microscopy. Chemicals and reagents for DNA manipulation were purchased from Takara Co. (Tokyo, Japan) and SolGent Co. (Daejeon, Korea). Chromosomal DNA purification for 16S rRNA sequencing was performed using the method of Berns and Thomas (1965). The DNA oligonucleotide primers were 27F (5'-AGAGTTTGATCCTGGCTCAG-3') and 1492R (5'-GGCTACCTTGTTACGACTT-3') and were synthesized by SolGent Co. (Dunbar et al., 2000). The reaction mixture (final volume, 50 μL) contained Taq polymerase 0.5 μL (2.5 U),
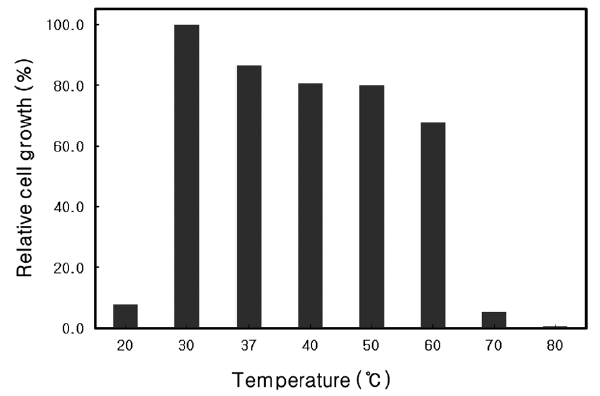
Nutrient broth (Difco Co.) with an additional 3% NaCl was used to culture the isolate. To determine the optimal temperature and pH for growth, the strain was cultured at various temperatures between 20 and 80℃ at pH 7, and at pH 5.0-10.0 at 37℃ . Cultures were incubated at 150 rpm for 24 h and the absorbance at 600 nm was measured.
The isolate was cultured in Nutrient broth (pH 6.0) containing 0-10% (w/v) NaCl at 30℃ for 24 h and the absorbance at 600 nm was measured.
>
Effect of agar concentration
The effect of agar concentration on growth of the isolate was determined. The isolate was cultured at 30℃ for 30 h at 150 rpm in Nutrient broth containing 0-0.4% (w/v) agar with 3% NaCl at pH 6.0 and the absorbance at 600 nm was measured every 3 h.
Thin-layer chromatography (TLC) of agar hydrolysate was performed on a Silica Gel 600 glass plate (F254; Merck Co., Darmstadt, Germany). To confirm the agarolytic activity of
>
Isolation of agarase-producing strain
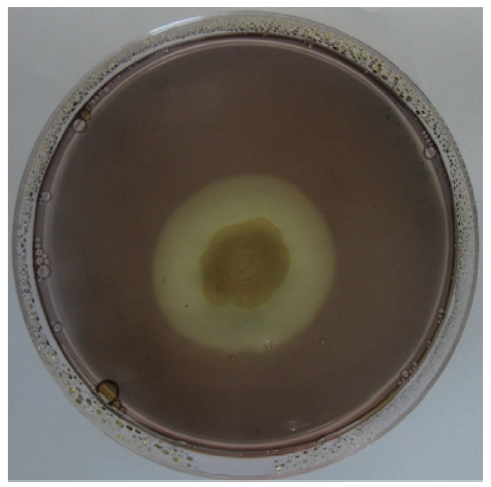
A bacterial strain that produced an extracellular agarolytic enzyme was isolated from seawater near Busan, Korea. Bacterial cells that can hydrolyze agar create a shallow depression or liquefaction around colonies on a 1.5% agar plate. Additionally, Lugol’s iodine solution was used to confirm the presence of haloes surrounding the colonies. Unstained haloes around colonies on agar plates treated with iodine show that the properties of the gel have been changed by the action of agarase diffusing out from the bacteria. The binding of iodine to agar gels to give a colored complex depends on the structure of the gel matrix. As shown in Fig. 1, the isolate produced extracellular agarase, resulting in a noticeable halo around the colony after staining with Lugol’s iodine solution.
>
Identification of the agarolytic strain
For taxonomic identification, the 16S rRNA sequence (1,457 bp) was determined and compared with sequences available in the GenBank database using the BLAST search program.
The genus Microbulbifer was first proposed by Gonzalez
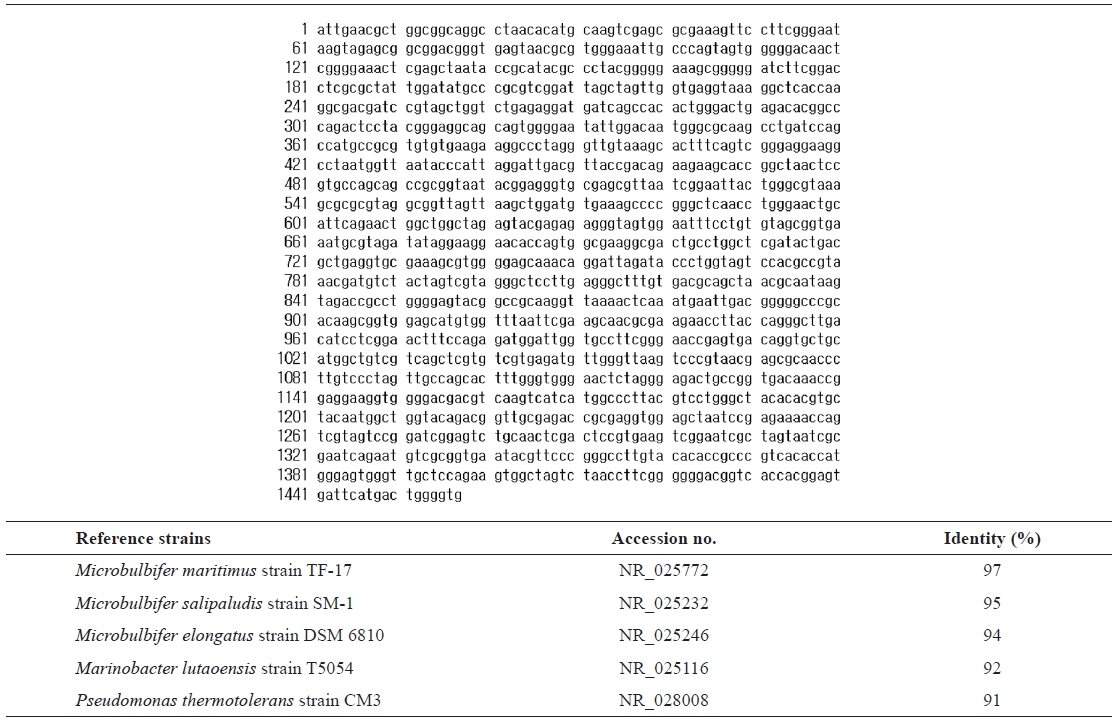
16s rRNA sequence (1457 bp) and identification of the isolated strain SD-1 by homology search based on 16S rRNA
which are typical properties of
The genus
et al. (1997) for
>
Optimal growth temperature of Microbulbifer sp. SD-1
The growth rate of
similar to that of
>
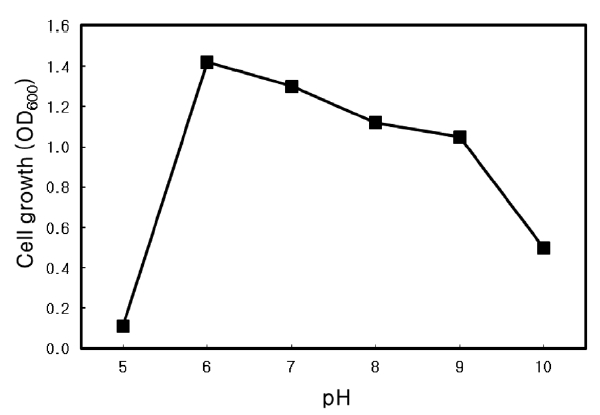
Optimal pH of Microbulbifer sp. SD-1
The optimal pH for growth of
>
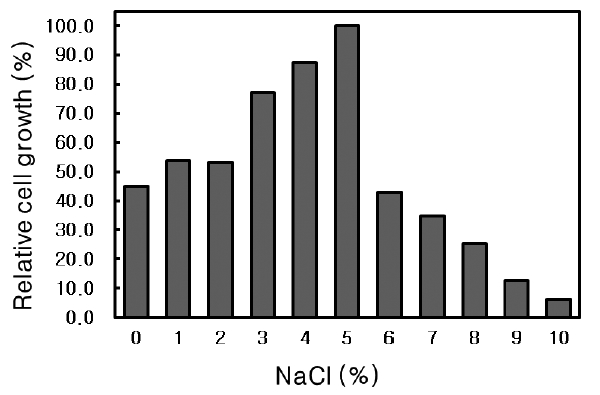
NaCl requirement of Microbulbifer sp. SD-1
The effect of NaCl concentration on the growth of
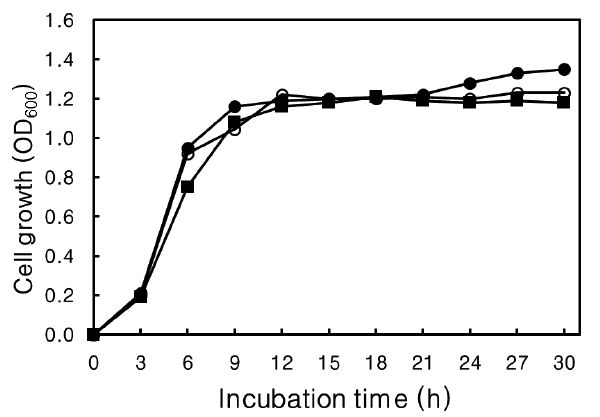
Agar, a type of polysaccharide, can be a source of carbohydrate for strains that can hydrolyze agar into smaller saccharides. The effect of agar on growth rate was determined (Fig. 6). No effect on growth rate was evident at more than 30 h culture or at the time to reach the stationary phase at 12 h, with or without agar in the medium. These results suggest that
>
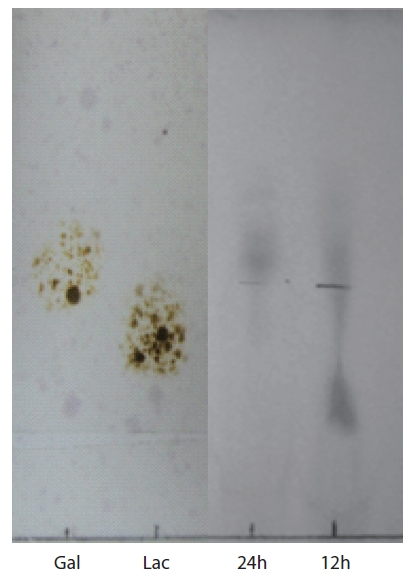
TLC analysis of agar hydrolysate
To confirm the agarolytic activity of