Karlodinium veneficum (Ballantine) J. Larsen, a mixotrophic dinoflagellate with a worldwide distribution,has been implicated in numerous fish kill events around the world (Place et al. 2008). K. veneficum produces water-soluble toxins that kill fish through gill disruption;however, researchers have successfully determined the structure of these karlotoxins (KmTxs), as they occur in K. veneficum (Kempton et al. 2002, Place et al. 2008). K.veneficum superficially resembles other small dinoflagellates< 15 ㎛ in size, including Pfiesteria piscicida and P. shumwayae, which commonly co-occur with K. veneficum in the marine environment (Litaker et al. 2005, Garces et al. 2006). These species are characterized by their distinct Kofoidian thecal plate formula. Definitive identification of these dinoflagellates relies on ultrastructural analyses via scanning electron microscopy (SEM), which are labor-intensive and unsuitable for rapid sample processing(Bergholtz et al. 2006, Garces et al. 2006). The real-time polymerase chain reaction (PCR) technique very sensitively detects and quantifies DNA over a broad dynamic range (Walker 2002). Certain studies have used this method for detecting and quantifying a number of dinoflagellates (Bowers et al. 2000, Park et al. 2009). A previous study developed the TaqMan and SYTO9 format real-time PCR probes for K. veneficum, and these probes showed comparable results and high detection sensitivities(Park et al. 2009). In this study, we used a TaqManbased real-time PCR probe that does not require a melting curve analysis to investigate temporal variations of K.veneficum abundances in Obido waters, Tongyeong, the location of many finfish farms.
We obtained Karlodinium veneficum (CCMP 415) from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton (CCMP). The culture was maintained in 30‰ f/2 medium (Guillard and Ryther 1962), without sodium silicate, at 20°C, under cool white fluorescent lamps (100-㎛ol photons m-2 s-1) on a 12 : 12-h light : dark cycle. Surface water temperature and salinity were measured in situ with a water quality monitor (YSI 600 XL; YSI Nanotech, Yellow Spring, OH, USA).
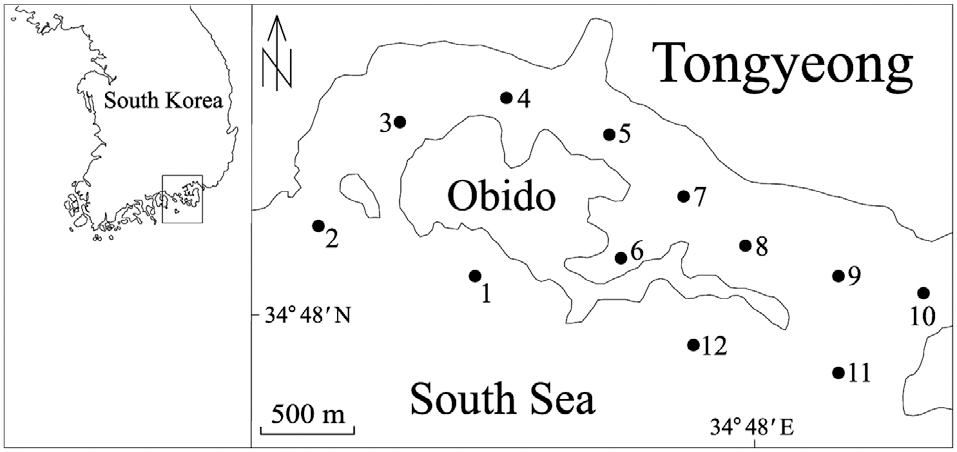
From April through December 2010, we collected 250 mL surface water samples, at 1 month intervals, from each of 12 stations in Obido, Tongyeong, Korea (Fig. 1). We filtered the water samples onto a 1.2 ㎛ pore, 25 mm diameter glass microfiber GF/C filter (Whatman Ltd., Maidstone, England), placed each filtered sample in a 2 mL microcentrifuge tube, and stored it at -70°C until DNA extraction. To prevent target DNA degradation, we processed these filtering samples upon a research vessel and extracted the samples genomic DNA from the samples within 2 months, using a phenol-chloroform extraction protocol (Hosoi-Tanabe and Sako 2005).
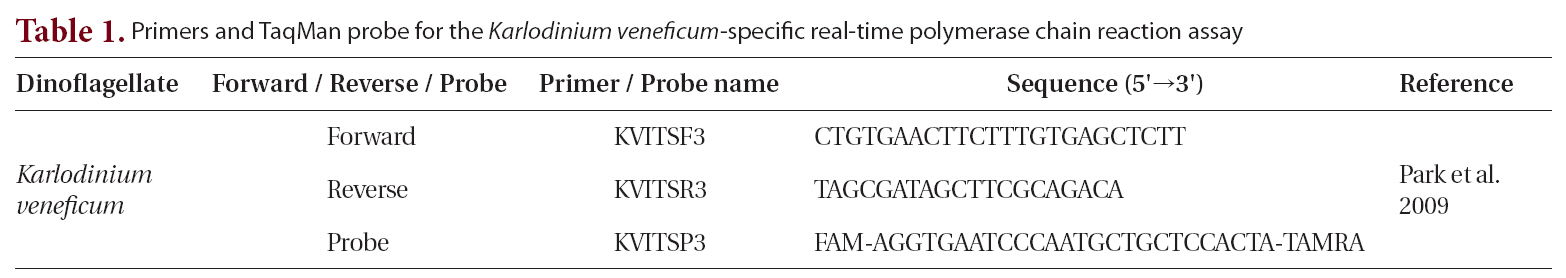
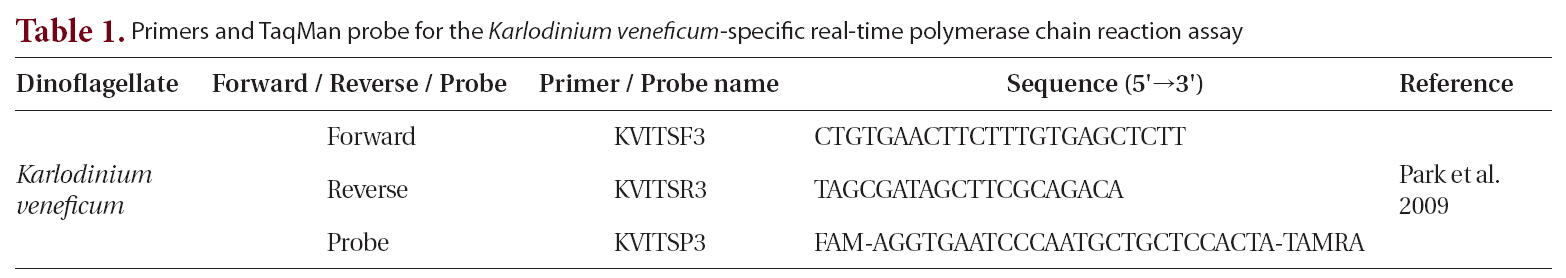
We used the following reagents to create the reaction mixture, to a final volume of 10 μL : 5 μL of platinum quantitative PCR supermix-UDG (Invitrogen, Eugene, OR, USA); forward and reverse primers, each at a final concentration of 0.3 μM; fluorogenic probe at a final concentration of 0.1 μM; 0.5 μL of template DNA (Park et al. 2009); and PCR grade water (Table 1). The thermal cycling condition comprised 2 min at 50°C and 2 min at
12
95°C, followed by 45 ten-s cycles at 95°C and 45 s at 60°C. All samples were analyzed in triplicate. We collected the fluorescence data at the end of each cycle, while a Rotor-Gene 6000 instrument (Corbett Research, Sydney, Australia) automatically determined the cycle threshold. To remove the surface waters’ PCR inhibitors, we diluted the template DNA tenfold before use.
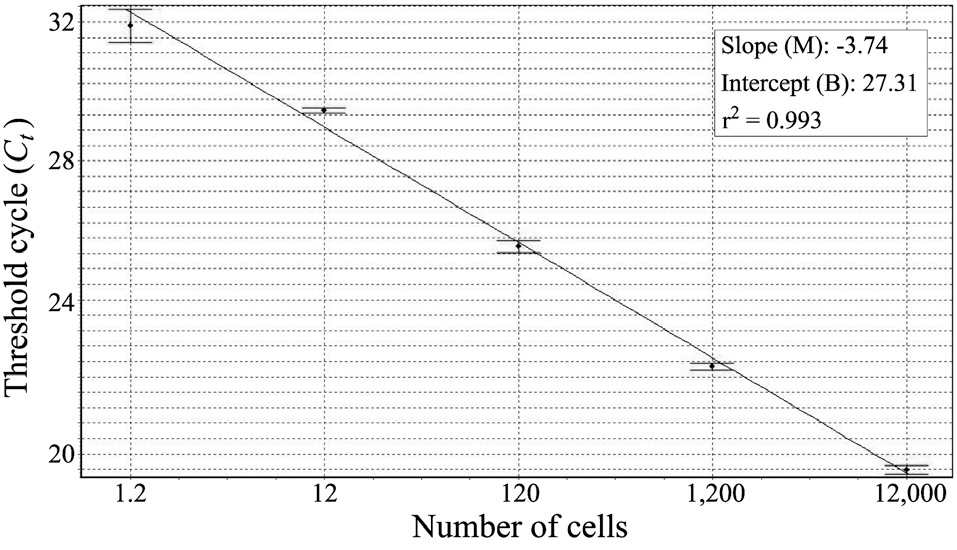
Before harvesting the cells (12,000 cells), we collected the laboratory-cultured K. veneficum and estimated its cell numbers via light microscopy (LM). After extracting the DNA, we used 10-fold serial dilutions of the DNA extracts to construct the standard curve (triplicate measurements by means of real-time PCR). To evaluate the success of the real-time PCR measurements, we calculated the correlation coefficient (r2), reaction efficiency [-1 + 10(-1/slope)], slope (M), and intercept (B), using the Rotor-Gene software version 1.7 (build 61; Corbett Research, Cambridge, UK). In the environmental samples, we calculated the target species’ cell numbers as Ct values and measured them via comparison with the standard curve. In addition, we observed the surface water samples via LM to check the K. veneficum cell numbers.
We constructed a standard curve with known cell numbers and established a strong linear relationship between the Ct and the log of the starting cell number, with a correlation coefficient (r2) ≥ 0.993 (Fig. 2). The assay could detect less than one K. veneficum cell in a reaction.
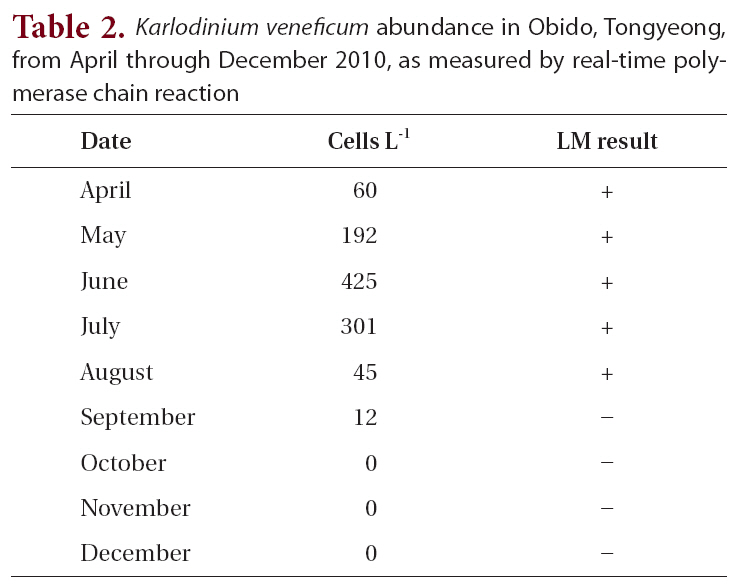
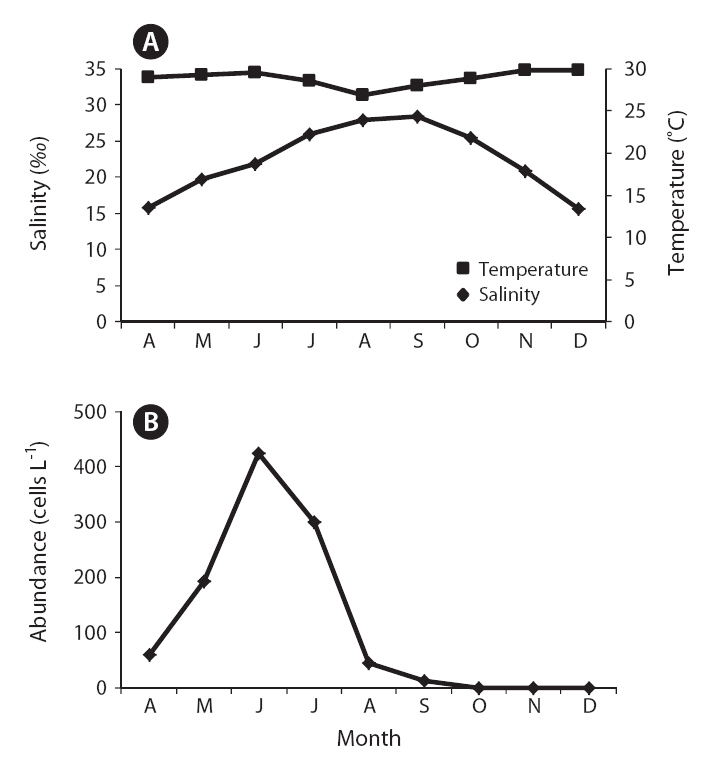
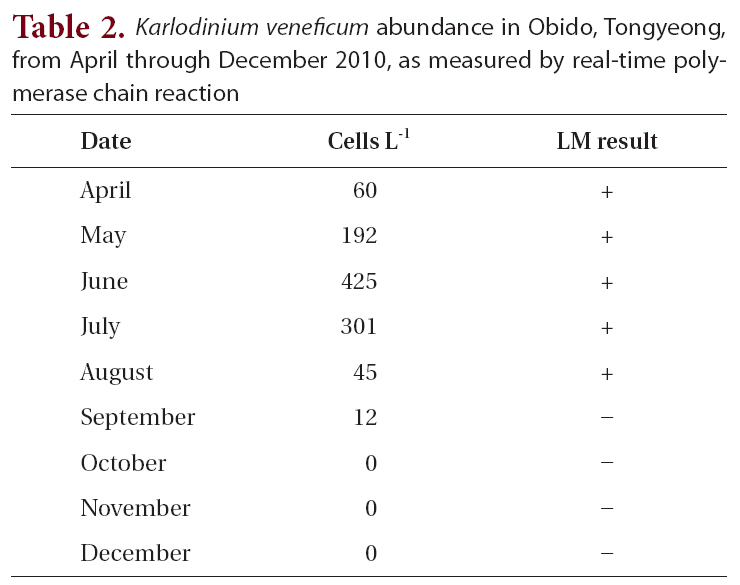
The field survey of the Obido waters showed that K. veneficum cell concentrations were generally low (Table 2, Fig. 3). The 12 sampling stations’ average cell numbers ranged from 12 to 425 cells L-1, and these cell numbers increased to 425 cells L-1 in June. K. veneficum was more abundant in early summer in the Obido area. The microscopic analysis revealed low K. veneficum and Karlodinium spp. cell abundances during the survey, and we observed these species only in samples that tested positive via the real-time PCR assay (Table 2). Surface water temperature and salinity ranged from 13.4-24.3°C and 31.3-34.8‰, respectively (Fig. 3).
Researchers have used a number of molecular methods, including real-time PCR, fluorescent in situ hybridization (FISH), sandwich hybridization, competitive PCR, a heteroduplex mobility assay, and fluorescent fragment PCR, to identify dinoflagellates rapidly (Oldach et al. 2000, Haywood et al. 2007, Park et al. 2007). TaqMan based real-time PCR probes have been previously designed to the internal transcribed spacer (ITS) rDNA and small subunit rDNA for the quantitative detection of K. veneficum (Handy et al. 2008, Park et al. 2009). Studies have extensively tested those species-specific assays against related organisms for their assay specificity and have successfully used these assays specifically for detecting K. veneficum in environmental samples (Handy et al. 2008, Park et al. 2009). The real-time PCR assay this study used specifically and sensitively detected K. veneficum, allowing us to identify low quantities of this dinoflagellate in our water samples (below one cell per reaction;
the real-time PCR could theoretically detect even one copy of the dinoflagellate’s DNA). The rDNA-based real-time PCR probe possesses such a high sensitivity because the rDNA in most eukaryotes is repeated in tandem at a high copy number. For example, one study reports 100 to 200 copies in one P. piscicida cell (Saito et al. 2002).
K. veneficum commonly co-occurs with morphologically similar species, such as Pfiesteria species, and research has correlated K. veneficum with numerous fish kill events in many countries (Fensin 2006, Place et al. 2008). Small gymnodinioid dinoflagellates, including K. veneficum, have been associated with toxic activity since the 1950s (Ballantine 1956, Place et al. 2008). This species produces a suite of compounds with hemolytic, cytotoxic, and ichthyotoxic properties (Deeds et al. 2002) which are often known as KmTxs. In laboratory bioassays, the purified toxins cause the deaths of supportive cells in menhaden gills and eventually result in fish mortality (Deeds et al. 2006). In the present study, a temporal survey of K. veneficum cell densities in Obido waters shows K. veneficum (maximum 425 cells L-1) mostly peaked in June, indicating potential, early-summer KmTx fish kills in the Obido area (16.9-22.3°C and 33.4-34.5‰) if blooms occur during favorable environmental conditions. In K. veneficum cells, the KmTxs’ toxicities are KvTX1, 11.6 ± 5.4 ng mL-1 and KvTX2, 47.7 ± 4.2 ng mL-1 at 4.0 × 106 cells L-1 (Galimany et al. 2008). Researchers have generally reported that fish kill events in nature correlate with this species when it is at over 300 cells mL-1. Although studies have also reported negative effects on marine animals, such as mussels, at pre-bloom concentrations (Galimany et al. 2008), the cell amounts that this study found might have negligible effects on aquaculture animals such as oysters, mussels, and finfishes in Obido waters.
In conclusion, the real-time PCR assay was highly sensitive and specific for detecting and quantifying K. veneficum in the environment, and we successfully implemented this real-time PCR assay in a distributional study of this species in the Obido area. The results of this field survey via real-time PCR suggests K. veneficum occurs commonly in the warmer season in the Obido area and that researchers need to make ongoing investigations of the relationship between environmental factors and cell occurrences to illuminate the bloom dynamics of K. veneficum.